Spatial Data - Displaying time series, spatial and space-time data with R
Table of Contents
Code
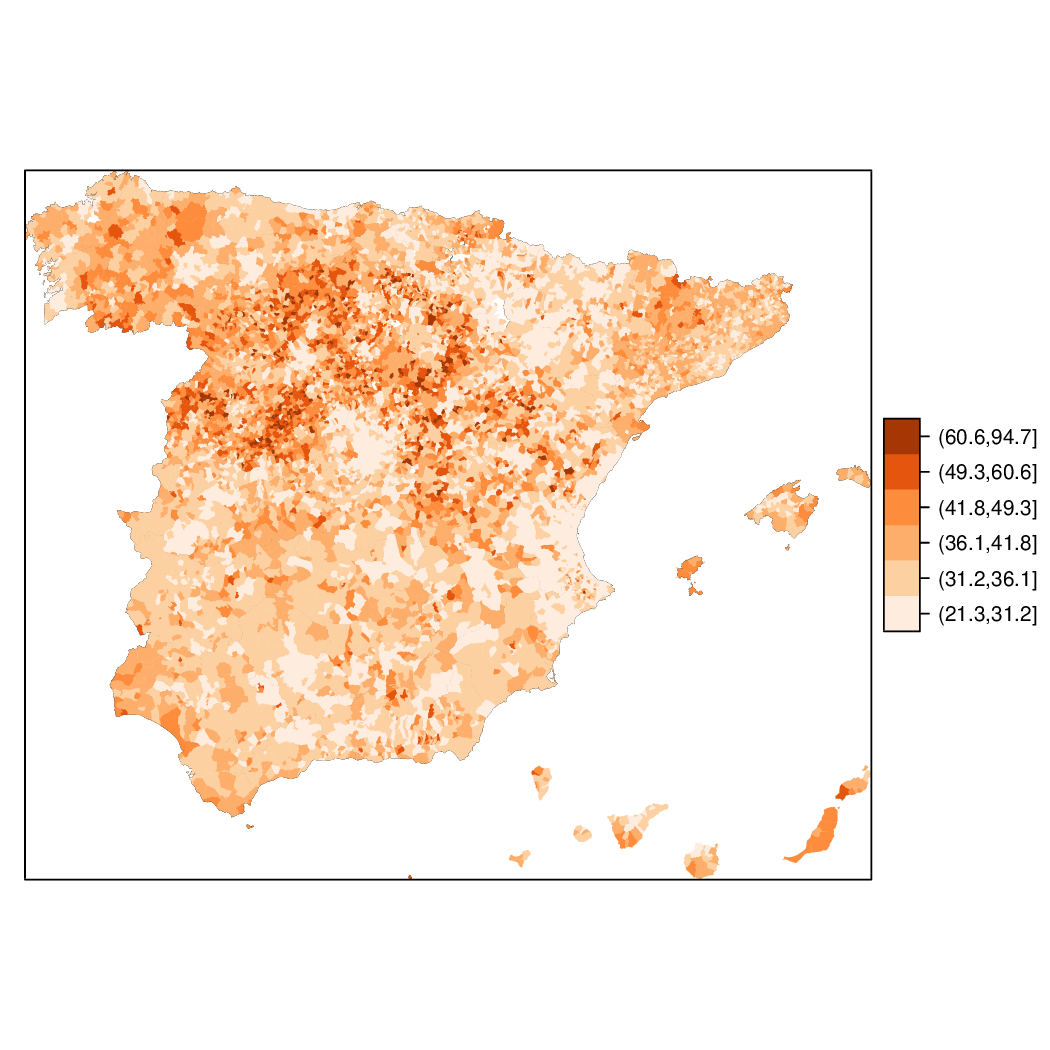
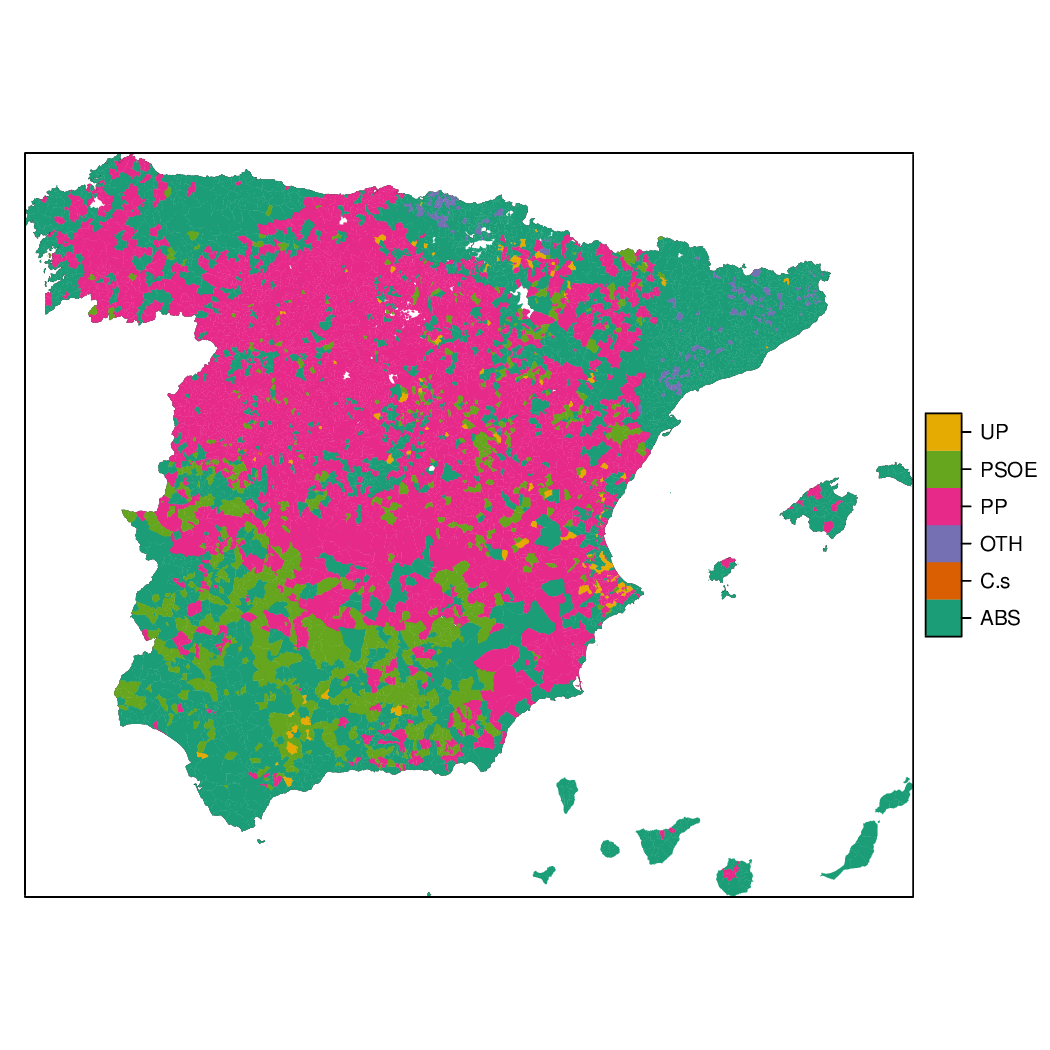
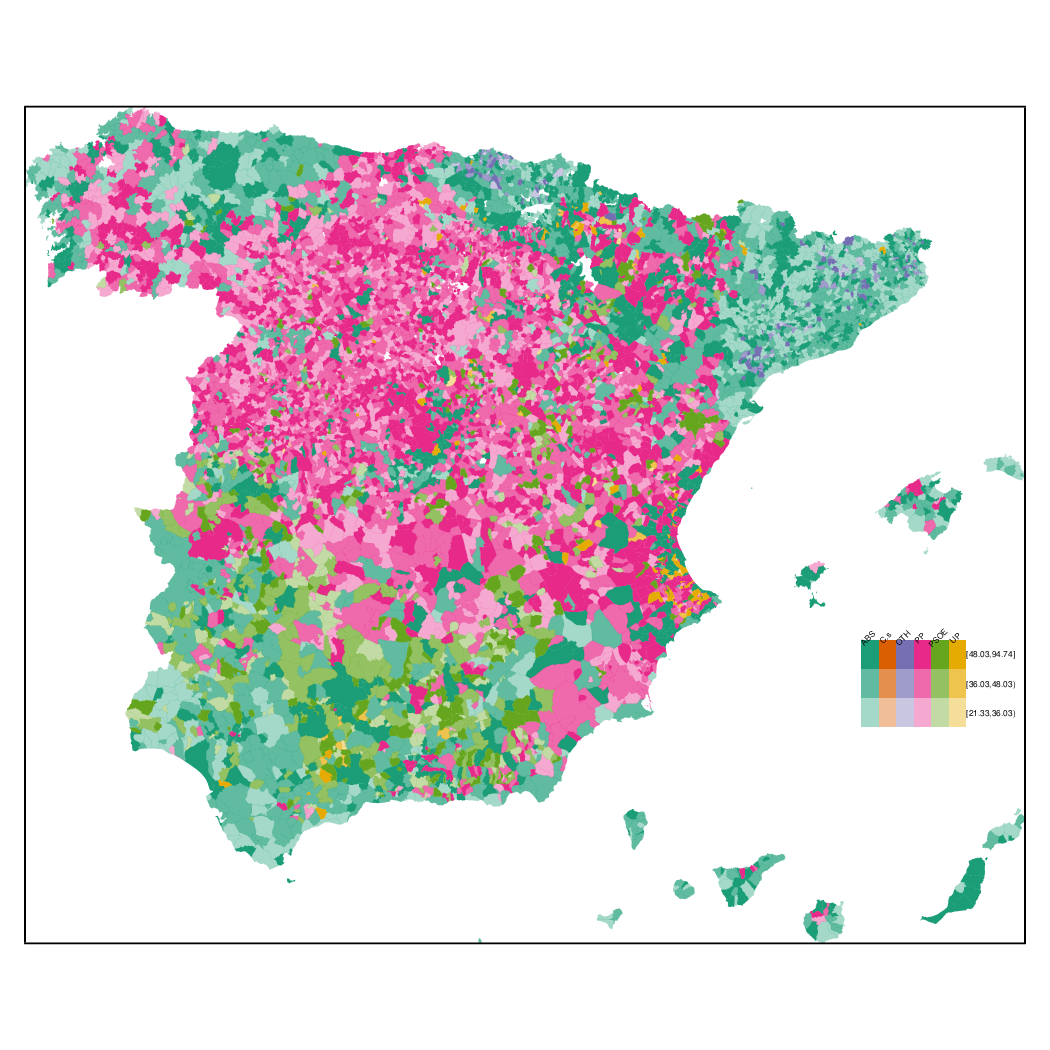
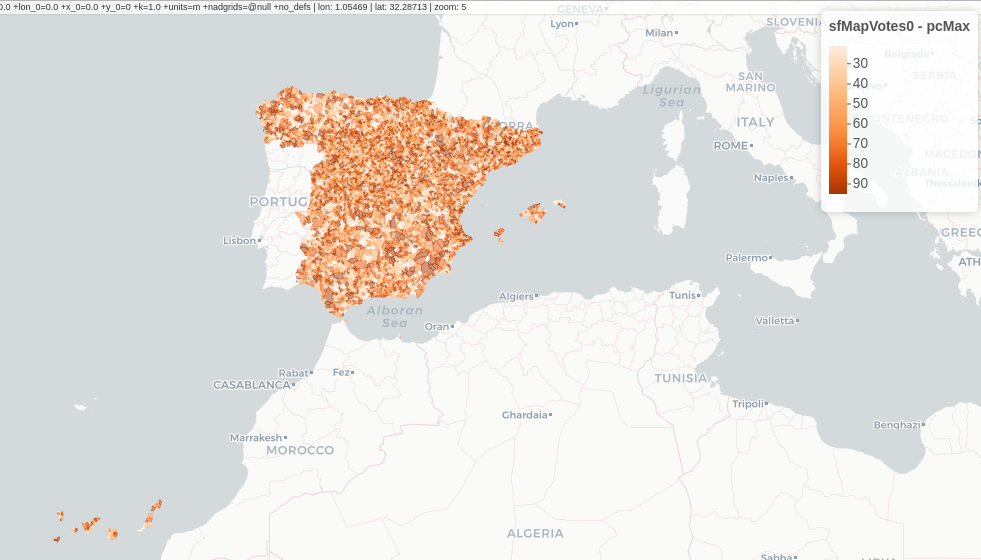
Choropleth maps
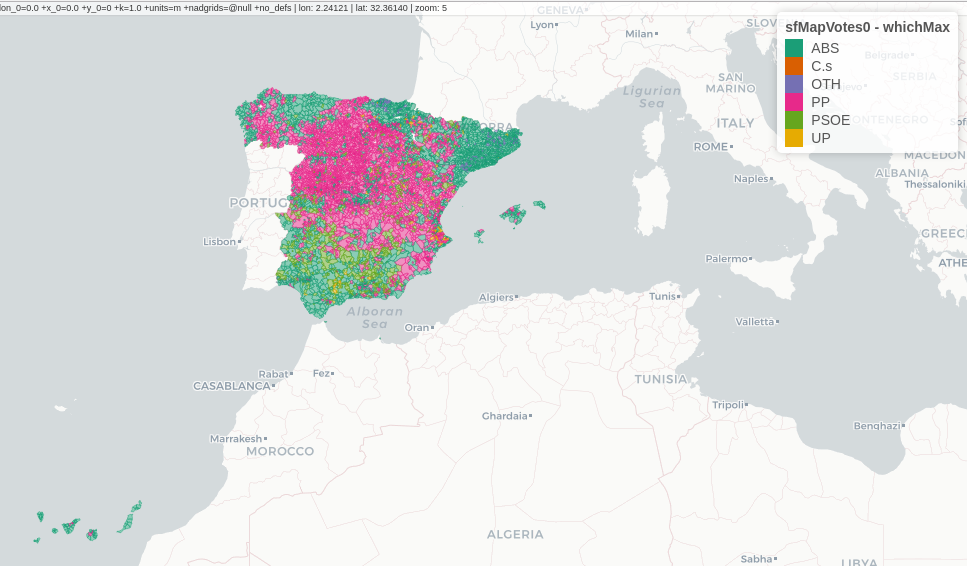
################################################################## ## Initial configuration ################################################################## ## Clone or download the repository and set the working directory ## with setwd to the folder where the repository is located. library(lattice) library(ggplot2) ## latticeExtra must be loaded after ggplot2 to prevent masking of its ## `layer` function. library(latticeExtra) source('configLattice.R') ################################################################## ################################################################## ## Read data ################################################################## ## sp approach library(sp) library(rgdal) spMapVotes <- readOGR("data/spMapVotes.shp", p4s = "+proj=utm +zone=30 +ellps=GRS80 +units=m +no_defs") ## sf approach library(sf) sfMapVotes <- st_read("data/spMapVotes.shp") st_crs(sfMapVotes) <- 25830 ################################################################## ## Province Boundaries ################################################################## ## sp provinces <- readOGR("data/spain_provinces.shp", p4s = "+proj=utm +zone=30 +ellps=GRS80 +units=m +no_defs") provinceLines <- list("sp.polygons", provinces, lwd = 0.1) ## sf sfProvs <- st_read("data/spain_provinces.shp") st_crs(sfProvs) <- 25830 ################################################################## ## Quantitative variable ################################################################## library(RColorBrewer) ## Number of intervals (colors) N <- 6 ## Sequential palette quantPal <- brewer.pal(n = N, "Oranges") ## Number of cuts ucN <- 1000 ## Palette created with interpolation ucQuantPal <- colorRampPalette(quantPal)(ucN) ## The polygons boundaries are not displayed thanks to col = 'transparent' spplot(spMapVotes["pcMax"], col.regions = ucQuantPal, cuts = ucN, ## Do not draw municipality boundaries col = 'transparent', ## Overlay province boundaries sp.layout = provinceLines) ################################################################## ## Data classification ################################################################## ggplot(as.data.frame(spMapVotes), aes(pcMax, fill = whichMax, colour = whichMax)) + geom_density(alpha = 0.1) + theme_bw() library(classInt) ## Compute intervals with the same number of elements intQuant <- classIntervals(spMapVotes$pcMax, n = N, style = "quantile") ## Compute intervals with the natural breaks algorithm intFisher <- classIntervals(spMapVotes$pcMax, n = N, style = "fisher") plot(intQuant, pal = quantPal, main = "") plot(intFisher, pal = quantPal, main = "") ## spplot solution ## Add a new categorical variable with cut, using the computed breaks spMapVotes$pcMaxInt <- cut(spMapVotes$pcMax, breaks = intFisher$brks) spplot(spMapVotes["pcMaxInt"], col = 'transparent', col.regions = quantPal, sp.layout = provinceLines) ## sf and geom_sf sfMapVotes$pcMaxInt <- cut(sfMapVotes$pcMax, breaks = intFisher$brks) ggplot(sfMapVotes) + ## Display the pcMaxInt variable... geom_sf(aes(fill = pcMaxInt), ## without drawing municipality boundaries color = "transparent") + scale_fill_brewer(palette = "Oranges") + ## And overlay provinces boundaries geom_sf(data = sfProvs, fill = 'transparent', ## but do not include them in the legend show.legend = FALSE) + theme_bw() ################################################################## ## Qualitative variable ################################################################## classes <- levels(factor(spMapVotes$whichMax)) nClasses <- length(classes) qualPal <- brewer.pal(nClasses, "Dark2") ## spplot solution spplot(spMapVotes["whichMax"], col.regions = qualPal, col = 'transparent', sp.layout = provinceLines) ## geom_sf solution ggplot(sfMapVotes) + geom_sf(aes(fill = whichMax), color = "transparent") + scale_fill_brewer(palette = 'Dark2') + geom_sf(data = sfProvs, fill = 'transparent', show.legend = FALSE) + theme_bw() ################################################################## ## Small multiples ################################################################## ## spplot version spplot(spMapVotes, "pcMaxInt", ## Formula to define the faceting: a panel for each level of ## whichMax formula = pcMaxInt ~ xlabelpoint + ylabelpoint | whichMax, col.regions = quantPal, col = 'transparent', sp.layout = provinceLines) ## ggplot2 version ggplot(sfMapVotes) + geom_sf(aes(fill = pcMaxInt), color = "transparent") + ## Define the faceting using two rows facet_wrap(~whichMax, nrow = 2) + scale_fill_brewer(palette = "Oranges") + geom_sf(data = sfProvs, fill = 'transparent', size = 0.1, show.legend = FALSE) + theme_bw() ################################################################## ## Bivariate map ################################################################## ## Number of intervals. N <- 3 ## Loop to create a bidimensional palette multiPal <- sapply(1:nClasses, function(i) { colorAlpha <- adjustcolor(qualPal[i], alpha = 0.4) colorRampPalette(c(qualPal[i], colorAlpha), alpha = TRUE)(N) }) ## Define the intervals intFisher <- classIntervals(spMapVotes$pcMax, n = N, style = "fisher") ## ... and create a categorical variable with them spMapVotes$pcMaxInt <- cut(spMapVotes$pcMax, breaks = intFisher$brks) pList <- lapply(1:nClasses, function(i){ ## Only those polygons corresponding to a level are selected mapClass <- subset(spMapVotes, whichMax == classes[i]) ## Palette pal <- multiPal[, i] ## Produce the graphic pClass <- spplot(mapClass, "pcMaxInt", col.regions = pal, col = 'transparent', colorkey = FALSE) }) names(pList) <- classes p <- Reduce('+', pList) op <- options(digits = 4) tabFisher <- print(intFisher) intervals <- names(tabFisher) options(op) library(grid) legend <- layer( { ## Position of the legend x0 <- 1000000 y0 <- 4200000 ## Width of the legend w <- 120000 ## Height of the legend h <- 100000 ## Colors grid.raster(multiPal, interpolate = FALSE, x = unit(x0, "native"), y = unit(y0, "native"), width = unit(w, "native"), height = unit(h, "native")) ## x-axis (qualitative variable) grid.text(classes, x = unit(seq(x0 - w * (nClasses -1)/(2*nClasses), x0 + w * (nClasses -1)/(2*nClasses), length = nClasses), "native"), y = unit(y0 + h/2, "native"), just = "bottom", rot = 10, gp = gpar(fontsize = 4)) ## y-axis (quantitative variable) Ni <- length(intervals) grid.text(intervals, x = unit(x0 + w/2, "native"), y = unit(seq(y0 - h * (Ni -1)/(2*Ni), y0 + h * (Ni -1)/(2*Ni), length = Ni), "native"), just = "left", gp = gpar(fontsize = 6)) }) ## Main plot p + legend ################################################################## ## Interactive Graphics ################################################################## library(mapview) sfMapVotes0 <- st_read("data/spMapVotes0.shp") st_crs(sfMapVotes0) <- 25830 ## Quantitative variable, pcMax mapView(sfMapVotes0, zcol = "pcMax", ## Choose the variable to display legend = TRUE, col.regions = quantPal) ## Qualitative variable, whichMax mapView(sfMapVotes0, zcol = "whichMax", legend = TRUE, col.regions = qualPal)
Raster maps
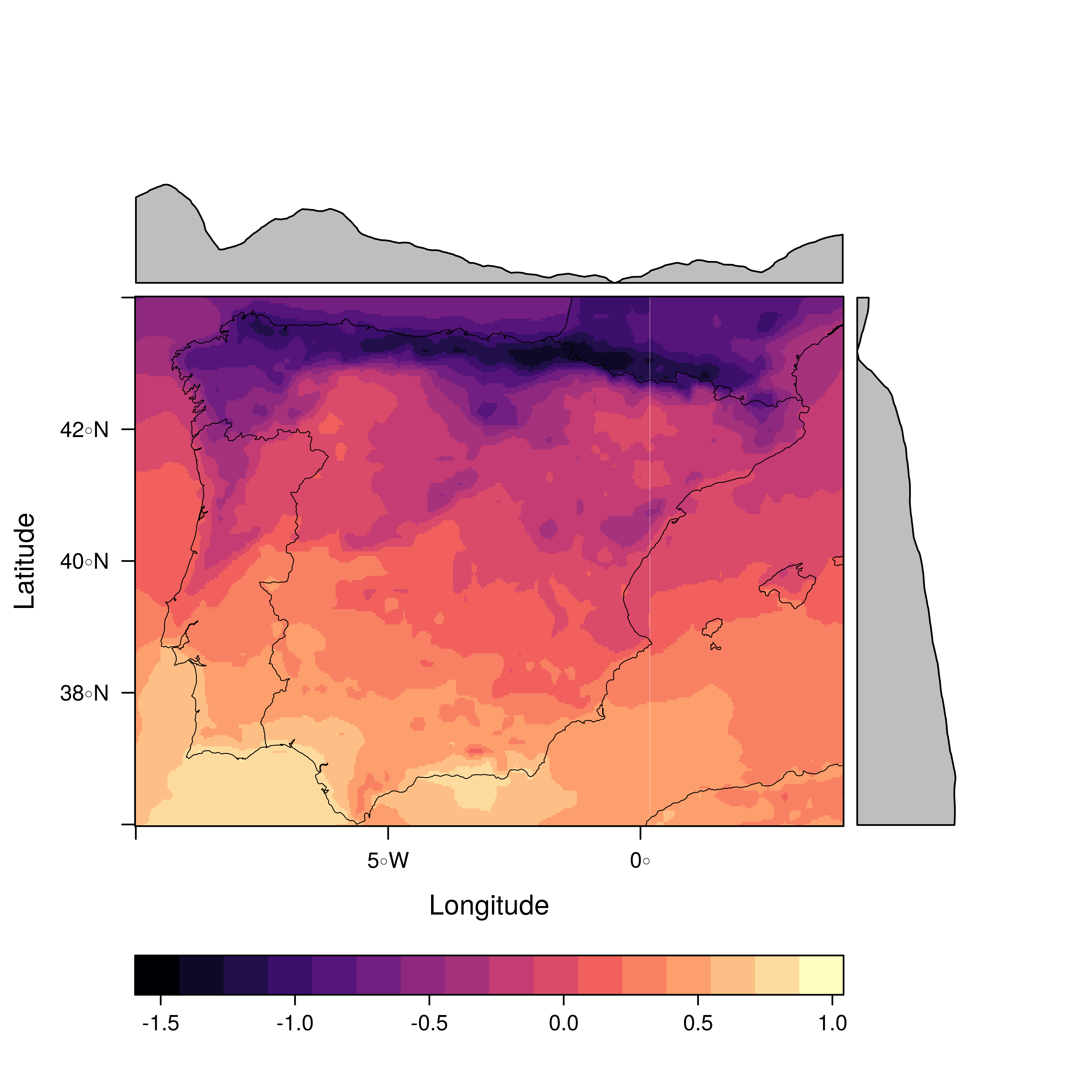
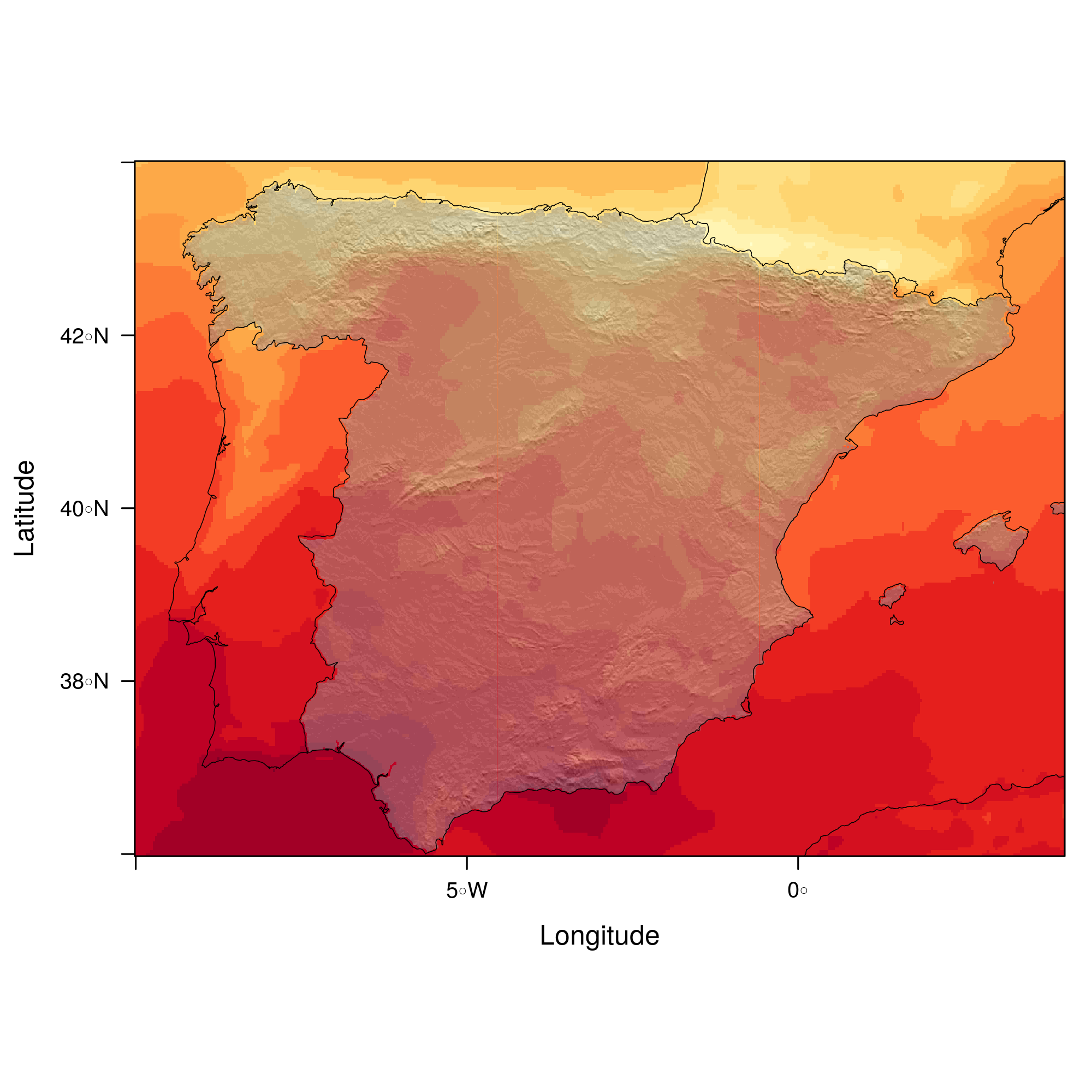
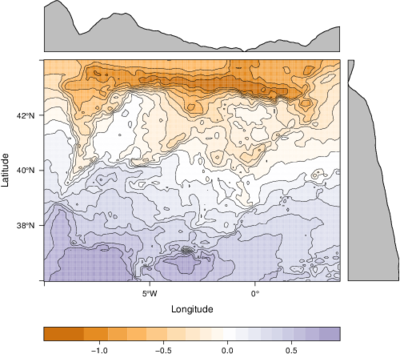
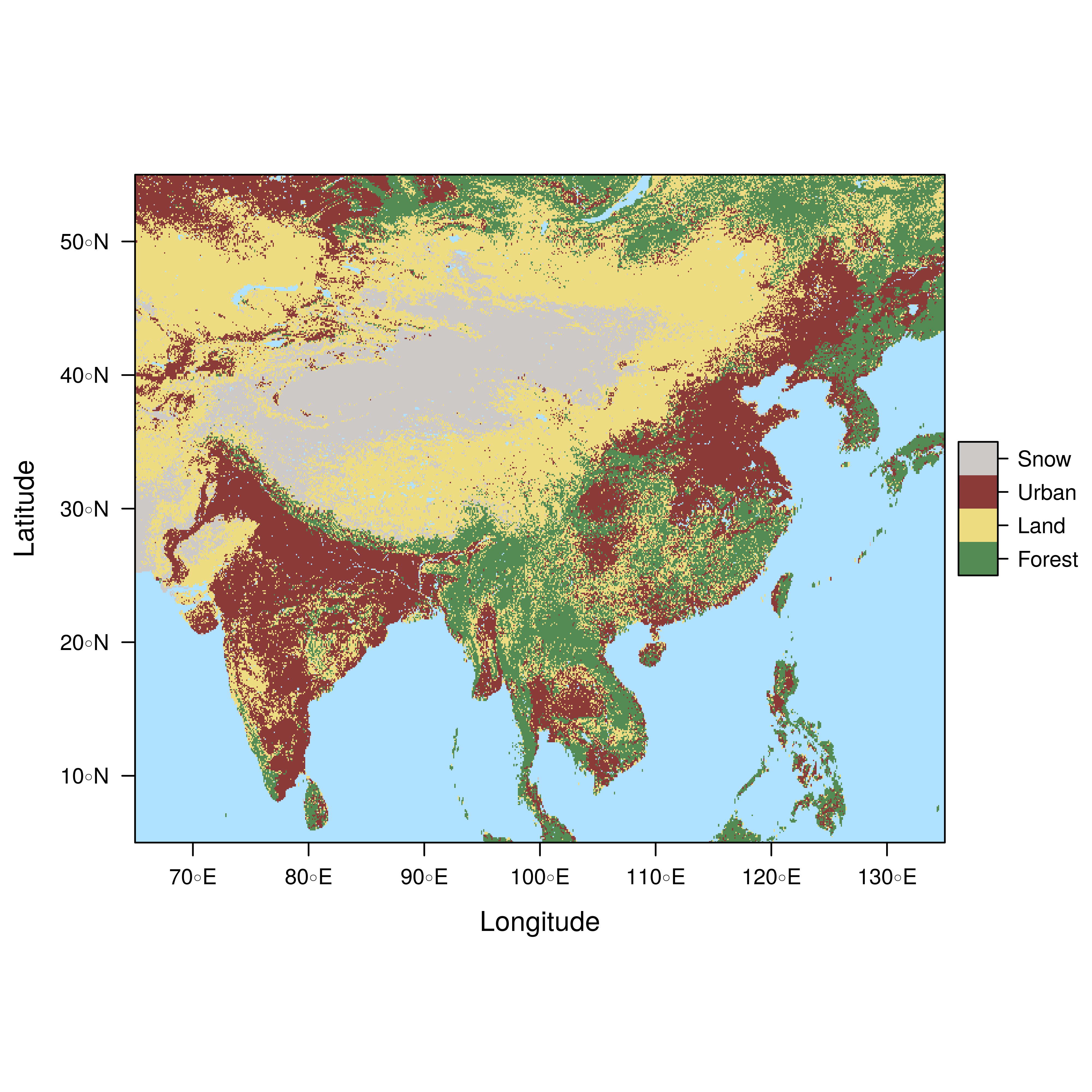
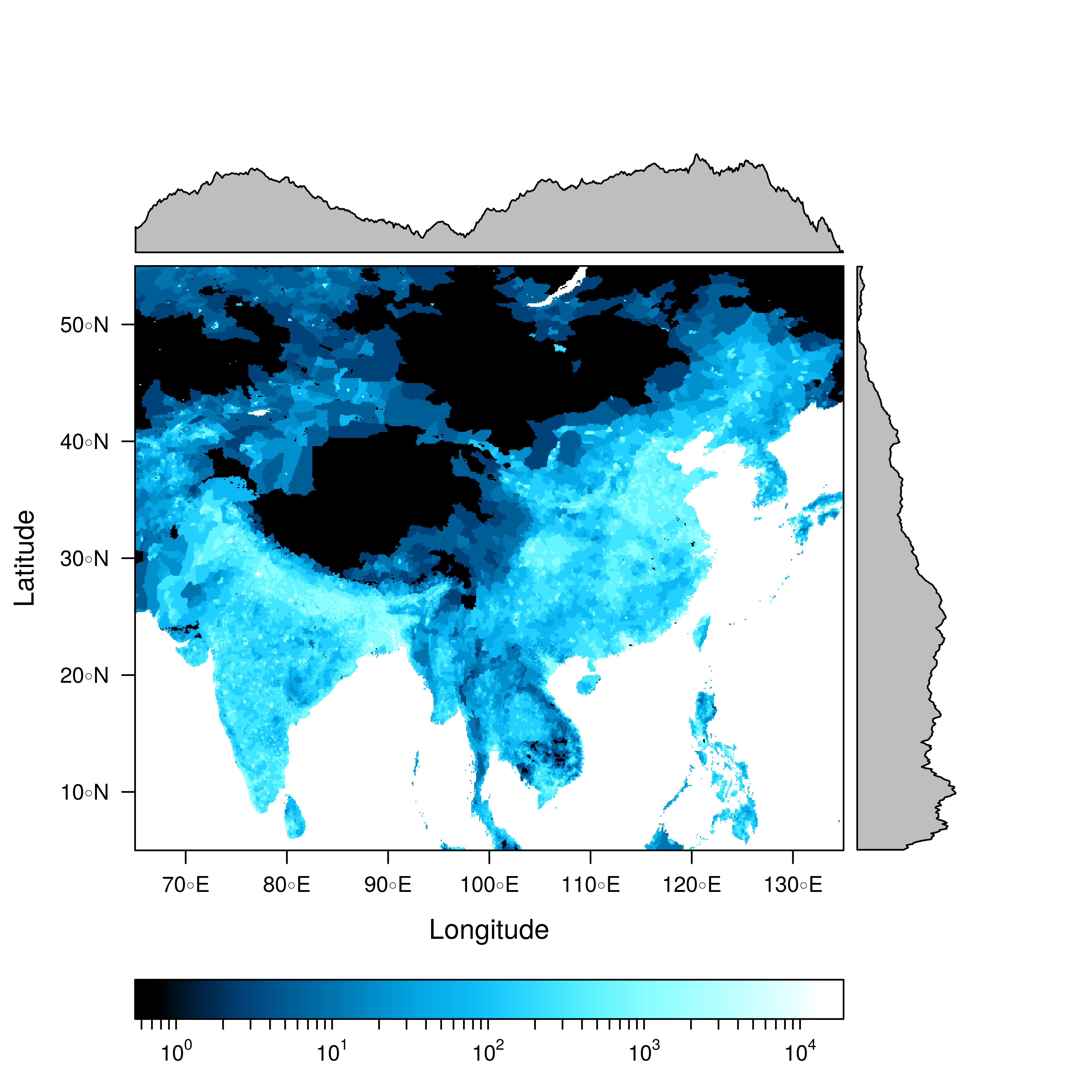
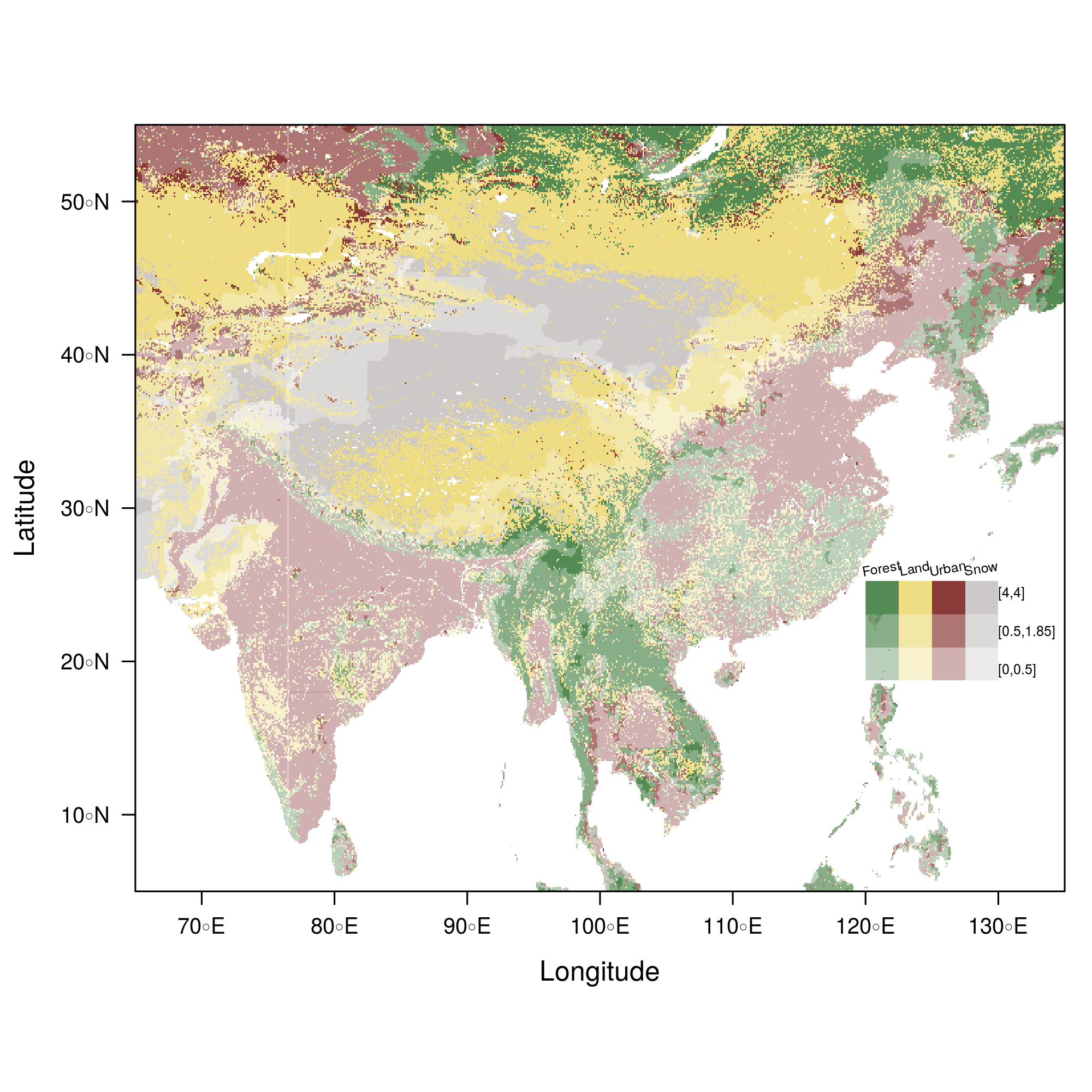
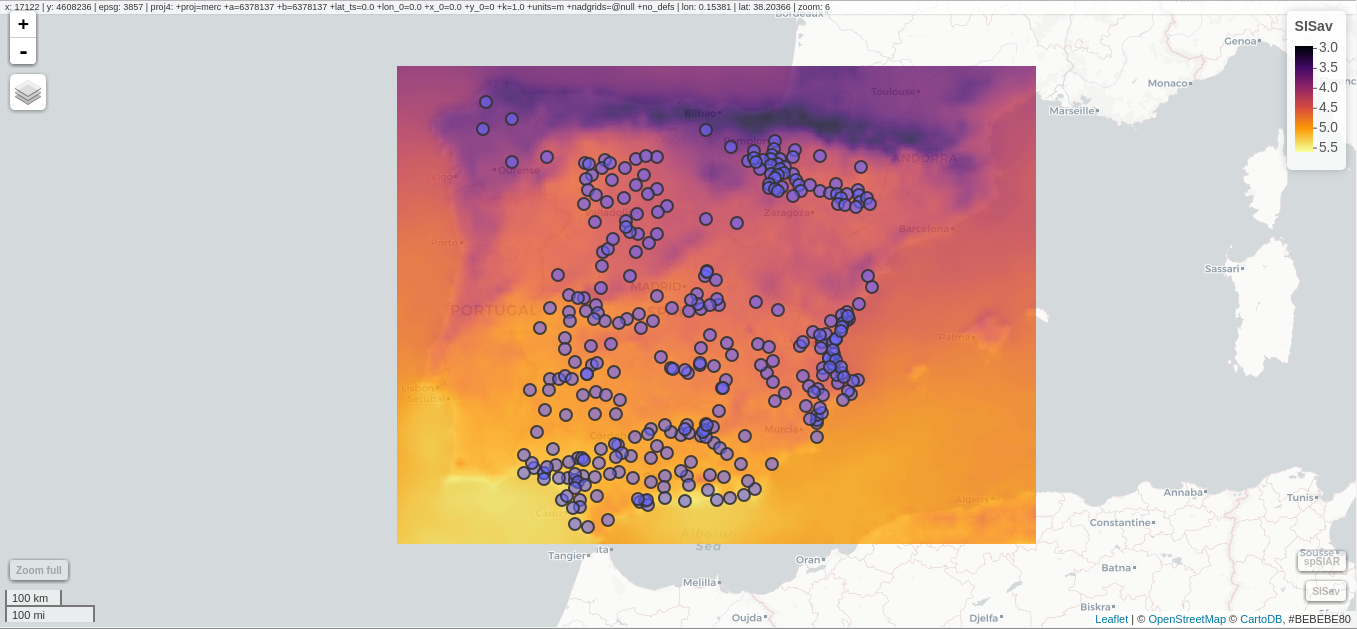
################################################################## ## Initial configuration ################################################################## ## Clone or download the repository and set the working directory ## with setwd to the folder where the repository is located. ################################################################## ## Quantitative data ################################################################## library(raster) library(rasterVis) SISav <- raster('data/SISav') levelplot(SISav) library(maps) library(mapdata) library(maptools) ## Extent of the Raster object ext <- as.vector(extent(SISav)) ## Retrieve the boundaries restricted to this extent boundaries <- map('worldHires', xlim = ext[1:2], ylim = ext[3:4], plot = FALSE) ## Convert the result to a SpatialLines object with the projection of ## the Raster object boundaries <- map2SpatialLines(boundaries, proj4string = CRS(projection(SISav))) ## Display the Raster object ... levelplot(SISav) + ## ... and overlay the SpatialLines object layer(sp.lines(boundaries, lwd = 0.5)) ################################################################## ## Hill shading ################################################################## old <- setwd(tempdir()) download.file('http://biogeo.ucdavis.edu/data/diva/msk_alt/ESP_msk_alt.zip', 'ESP_msk_alt.zip') unzip('ESP_msk_alt.zip', exdir = '.') DEM <- raster('ESP_msk_alt') slope <- terrain(DEM, 'slope') aspect <- terrain(DEM, 'aspect') hs <- hillShade(slope = slope, aspect = aspect, angle = 60, direction = 45) setwd(old) ## hillShade theme: gray colors and semitransparency hsTheme <- GrTheme(regions = list(alpha = 0.5)) levelplot(SISav, par.settings = YlOrRdTheme, margin = FALSE, colorkey = FALSE) + ## Overlay the hill shade raster levelplot(hs, par.settings = hsTheme, maxpixels = 1e6) + ## and the countries boundaries layer(sp.lines(boundaries, lwd = 0.5)) ################################################################## ## Diverging palettes ################################################################## meanRad <- cellStats(SISav, 'mean') SISav <- SISav - meanRad xyplot(layer ~ y, data = SISav, groups = cut(x, 5), par.settings = rasterTheme(symbol = plinrain(n = 5, end = 200)), xlab = 'Latitude', ylab = 'Solar radiation (scaled)', auto.key = list(space = 'right', title = 'Longitude', cex.title = 1.3)) divPal <- brewer.pal(n = 9, 'PuOr') divPal[5] <- "#FFFFFF" showPal <- function(pal) { N <- length(pal) image(1:N, 1, as.matrix(1:N), col = pal, xlab = '', ylab = '', xaxt = "n", yaxt = "n", bty = "n") } showPal(divPal) divTheme <- rasterTheme(region = divPal) levelplot(SISav, contour = TRUE, par.settings = divTheme) rng <- range(SISav[]) ## Number of desired intervals nInt <- 15 ## Increment corresponding to the range and nInt inc0 <- diff(rng)/nInt ## Number of intervals from the negative extreme to zero n0 <- floor(abs(rng[1])/inc0) ## Update the increment adding 1/2 to position zero in the center of an interval inc <- abs(rng[1])/(n0 + 1/2) ## Number of intervals from zero to the positive extreme n1 <- ceiling((rng[2]/inc - 1/2) + 1) ## Collection of breaks breaks <- seq(rng[1], by = inc, length= n0 + 1 + n1) ## Midpoints computed with the median of each interval idx <- findInterval(SISav[], breaks, rightmost.closed = TRUE) mids <- tapply(SISav[], idx, median) ## Maximum of the absolute value both limits mx <- max(abs(breaks)) break2pal <- function(x, mx, pal){ ## x = mx gives y = 1 ## x = 0 gives y = 0.5 y <- 1/2*(x/mx + 1) rgb(pal(y), maxColorValue = 255) } ## Interpolating function that maps colors with [0, 1] ## rgb(divRamp(0.5), maxColorValue=255) gives "#FFFFFF" (white) divRamp <- colorRamp(divPal) ## Diverging palette where white is associated with the interval ## containing the zero pal <- break2pal(mids, mx, divRamp) showPal(pal) levelplot(SISav, par.settings = rasterTheme(region = pal), at = breaks, contour = TRUE) divTheme <- rasterTheme(regions = list(col = pal)) levelplot(SISav, par.settings = divTheme, at = breaks, contour = TRUE) library(classInt) cl <- classIntervals(SISav[], style = 'kmeans') breaks <- cl$brks ## Repeat the procedure previously exposed, using the 'breaks' vector ## computed with classIntervals idx <- findInterval(SISav[], breaks, rightmost.closed = TRUE) mids <- tapply(SISav[], idx, median) mx <- max(abs(breaks)) pal <- break2pal(mids, mx, divRamp) ## Modify the vector of colors in the 'divTheme' object divTheme$regions$col <- pal levelplot(SISav, par.settings = divTheme, at = breaks, contour = TRUE) ################################################################## ## Categorical data ################################################################## ## China and India ext <- extent(65, 135, 5, 55) pop <- raster('data/875430rgb-167772161.0.FLOAT.TIFF') pop <- crop(pop, ext) pop[pop==99999] <- NA landClass <- raster('data/241243rgb-167772161.0.TIFF') landClass <- crop(landClass, ext) landClass[landClass %in% c(0, 254)] <- NA ## Only four groups are needed: ## Forests: 1:5 ## Shrublands, etc: 6:11 ## Agricultural/Urban: 12:14 ## Snow: 15:16 landClass <- cut(landClass, c(0, 5, 11, 14, 16)) ## Add a Raster Attribute Table and define the raster as categorical data landClass <- ratify(landClass) ## Configure the RAT: first create a RAT data.frame using the ## levels method; second, set the values for each class (to be ## used by levelplot); third, assign this RAT to the raster ## using again levels rat <- levels(landClass)[[1]] rat$classes <- c('Forest', 'Land', 'Urban', 'Snow') levels(landClass) <- rat qualPal <- c('palegreen4', # Forest 'lightgoldenrod', # Land 'indianred4', # Urban 'snow3') # Snow qualTheme <- rasterTheme(region = qualPal, panel.background = list(col = 'lightskyblue1') ) levelplot(landClass, maxpixels = 3.5e5, par.settings = qualTheme) pPop <- levelplot(pop, zscaleLog = 10, par.settings = BTCTheme, maxpixels = 3.5e5) pPop ## Join the RasterLayer objects to create a RasterStack object. s <- stack(pop, landClass) names(s) <- c('pop', 'landClass') densityplot(~log10(pop), ## Represent the population groups = landClass, ## grouping by land classes data = s, ## Do not plot points below the curves plot.points = FALSE) ################################################################## ## Bivariate legend ################################################################## classes <- rat$classes nClasses <- length(classes) logPopAt <- c(0, 0.5, 1.85, 4) nIntervals <- length(logPopAt) - 1 multiPal <- sapply(1:nClasses, function(i) { colorAlpha <- adjustcolor(qualPal[i], alpha = 0.4) colorRampPalette(c(qualPal[i], colorAlpha), alpha = TRUE)(nIntervals) }) pList <- lapply(1:nClasses, function(i){ landSub <- landClass ## Those cells from a different land class are set to NA... landSub[!(landClass==i)] <- NA ## ... and the resulting raster masks the population raster popSub <- mask(pop, landSub) ## Palette pal <- multiPal[, i] pClass <- levelplot(log10(popSub), at = logPopAt, maxpixels = 3.5e5, col.regions = pal, colorkey = FALSE, margin = FALSE) }) p <- Reduce('+', pList) library(grid) legend <- layer( { ## Center of the legend (rectangle) x0 <- 125 y0 <- 22 ## Width and height of the legend w <- 10 h <- w / nClasses * nIntervals ## Legend grid.raster(multiPal, interpolate = FALSE, x = unit(x0, "native"), y = unit(y0, "native"), width = unit(w, "native")) ## Axes of the legend ## x-axis (qualitative variable) grid.text(classes, x = unit(seq(x0 - w * (nClasses -1)/(2*nClasses), x0 + w * (nClasses -1)/(2*nClasses), length = nClasses), "native"), y = unit(y0 + h/2, "native"), just = "bottom", rot = 10, gp = gpar(fontsize = 6)) ## y-axis (quantitative variable) yLabs <- paste0("[", paste(logPopAt[-nIntervals], logPopAt[-1], sep = ","), "]") grid.text(yLabs, x = unit(x0 + w/2, "native"), y = unit(seq(y0 - h * (nIntervals -1)/(2*nIntervals), y0 + h * (nIntervals -1)/(2*nIntervals), length = nIntervals), "native"), just = "left", gp = gpar(fontsize = 6)) }) p + legend ################################################################## ## 3D visualization ################################################################## plot3D(DEM, maxpixels = 5e4) ## Dimensions of the window in pixels par3d(viewport = c(0, 30, ## Coordinates of the lower left corner 250, 250)) ## Width and height writeWebGL(filename = 'docs/images/rgl/DEM.html', width = 800) ################################################################## ## mapview ################################################################## library(mapview) mvSIS <- mapview(SISav, legend = TRUE) SIAR <- read.csv("data/SIAR.csv") spSIAR <- SpatialPointsDataFrame(coords = SIAR[, c("lon", "lat")], data = SIAR, proj4str = CRS(projection(SISav))) mvSIAR <- mapview(spSIAR, label = spSIAR$Estacion) mvSIS + mvSIAR
Proportional symbol mapping
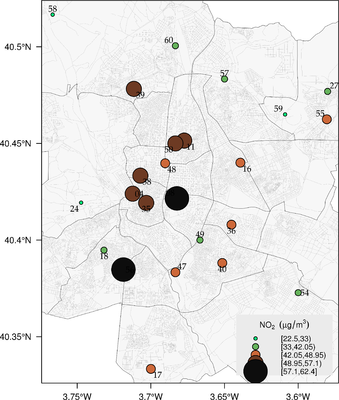
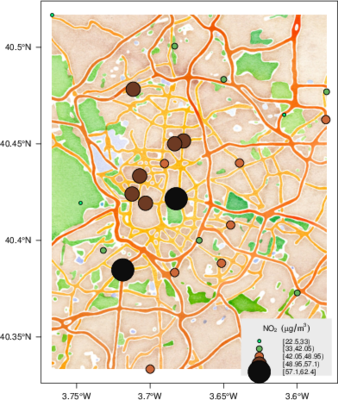
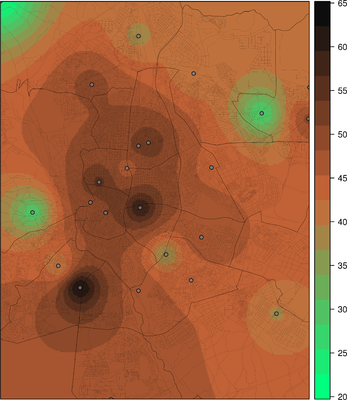
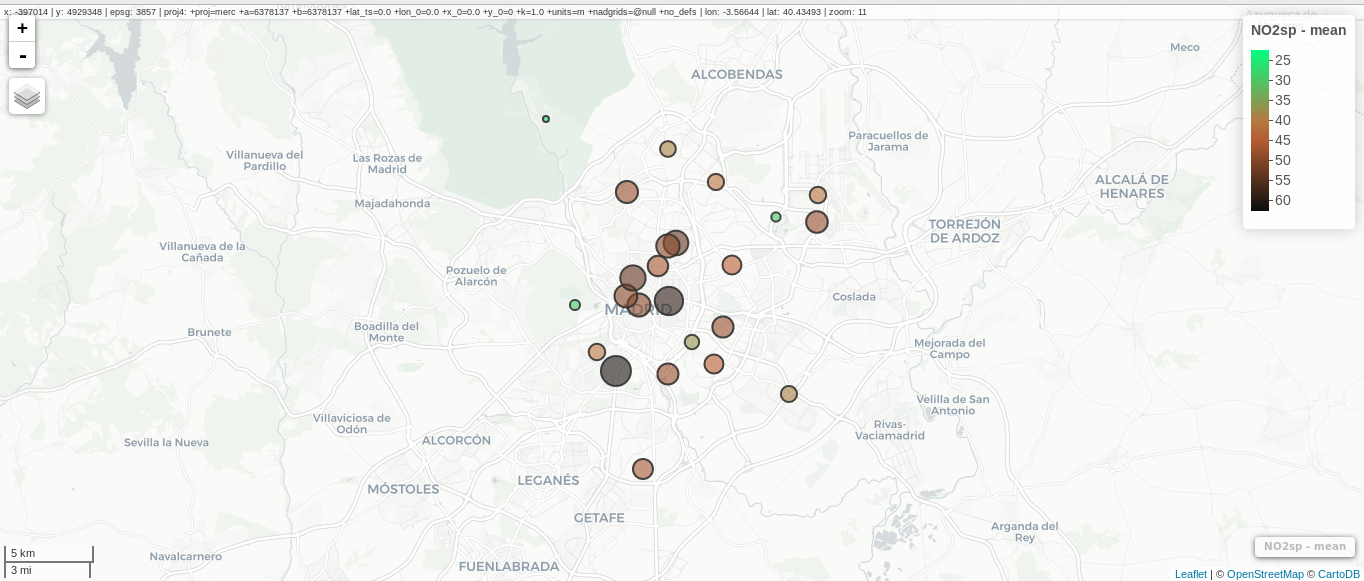
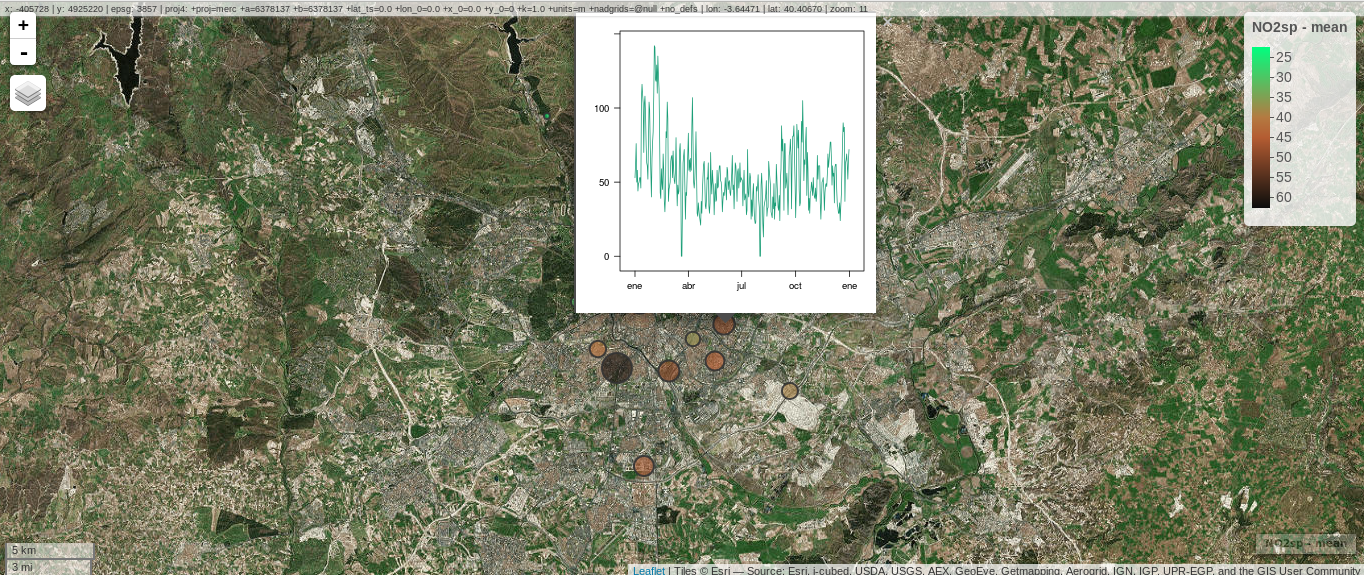
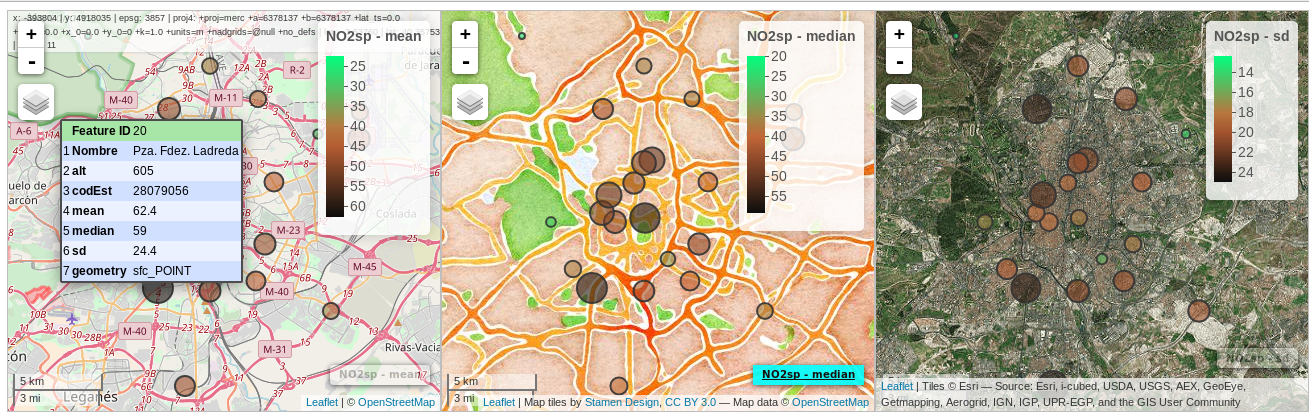
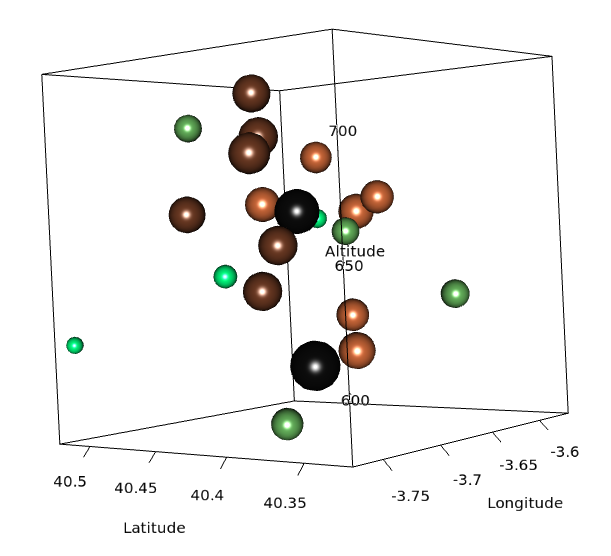
################################################################## ## Initial configuration ################################################################## ## Clone or download the repository and set the working directory ## with setwd to the folder where the repository is located. library(lattice) library(ggplot2) ## latticeExtra must be loaded after ggplot2 to prevent masking of its ## `layer` function. library(latticeExtra) source('configLattice.R') ################################################################## ################################################################## ## Proportional symbol with spplot ################################################################## library(sp) library(rgdal) NO2sp <- readOGR(dsn = 'data/', layer = 'NO2sp') airPal <- colorRampPalette(c('springgreen1', 'sienna3', 'gray5'))(5) spplot(NO2sp["mean"], col.regions = airPal, ## Palette cex = sqrt(1:5), ## Size of circles edge.col = 'black', ## Color of border scales = list(draw = TRUE), ## Draw scales key.space = 'right') ## Put legend on the right ################################################################## ## Proportional symbol with ggplot ################################################################## library(sf) NO2sf <- st_read(dsn = 'data', layer = 'NO2sp') ## Create a categorical variable NO2sf$Mean <- cut(NO2sf$mean, 5) ggplot(data = NO2sf) + geom_sf(aes(size = Mean, fill = Mean), pch = 21, col = 'black') + scale_fill_manual(values = airPal) + theme_bw() ################################################################## ## Optimal classification and sizes to improve discrimination ################################################################## library(classInt) ## The number of classes is chosen between the Sturges and the ## Scott rules. nClasses <- 5 intervals <- classIntervals(NO2sp$mean, n = nClasses, style = 'fisher') ## Number of classes is not always the same as the proposed number nClasses <- length(intervals$brks) - 1 op <- options(digits = 4) tab <- print(intervals) options(op) ## Complete Dent set of circle radii (mm) dent <- c(0.64, 1.14, 1.65, 2.79, 4.32, 6.22, 9.65, 12.95, 15.11) ## Subset for our dataset dentAQ <- dent[seq_len(nClasses)] ## Link Size and Class: findCols returns the class number of each ## point; cex is the vector of sizes for each data point idx <- findCols(intervals) cexNO2 <- dentAQ[idx] ## spplot version NO2sp$classNO2 <- factor(names(tab)[idx]) ## Definition of an improved key with title and background NO2key <- list(x = 0.99, y = 0.01, corner = c(1, 0), title = expression(NO[2]~~(paste(mu, plain(g))/m^3)), cex.title = 0.8, cex = 1, background = 'gray92') pNO2 <- spplot(NO2sp["classNO2"], col.regions = airPal, cex = dentAQ * 0.8, edge.col = 'black', scales = list(draw = TRUE), key.space = NO2key) pNO2 ## ggplot2 version NO2sf$classNO2 <- factor(names(tab)[idx]) ggplot(data = NO2sf) + geom_sf(aes(size = classNO2, fill = classNO2), pch = 21, col = 'black') + scale_fill_manual(values = airPal) + scale_size_manual(values = dentAQ * 2) + xlab("") + ylab("") + theme_bw() ################################################################## ## Spatial context with underlying layers and labels ################################################################## ################################################################## ## Static image ################################################################## ## Bounding box of data madridBox <- bbox(NO2sp) ## Extend the limits to get a slightly larger map madridBox <- t(apply(madridBox, 1, extendrange, f = 0.05)) library(ggmap) madridGG <- get_map(c(madridBox), maptype = 'watercolor', source = 'stamen') ## ggmap with ggplot NO2df <- as.data.frame(NO2sp) ggmap(madridGG) + geom_point(data = NO2df, aes(coords.x1, coords.x2, size = classNO2, fill = classNO2), pch = 21, col = 'black') + scale_fill_manual(values = airPal) + scale_size_manual(values = dentAQ*2) ## ggmap with spplot ## Project the data into the web mercator projection NO2merc <- spTransform(NO2sp, CRS("+init=epsg:3857")) ## sp.layout definition stamen <- list(panel.ggmap, ## Function that displays the object madridGG, ## Object to be displayed first = TRUE) ## This layout item will be drawn before ## the object displayed by spplot spplot(NO2merc["classNO2"], col.regions = airPal, cex = dentAQ * 0.8, edge.col = 'black', sp.layout = stamen, scales = list(draw = TRUE), key.space = NO2key) ################################################################## ## Vector data ################################################################## ################################################################## ## rgdal and spplot ################################################################## library(rgdal) ## nomecalles http://www.madrid.org/nomecalles/Callejero_madrid.icm ## Form at http://www.madrid.org/nomecalles/DescargaBDTCorte.icm ## Madrid districts unzip('Distritos de Madrid.zip') distritosMadrid <- readOGR('Distritos de Madrid/200001331.shp', p4s = '+proj=utm +zone=30') distritosMadrid <- spTransform(distritosMadrid, CRS = CRS("+proj=longlat +ellps=WGS84")) ## Madrid streets unzip('Callejero_ Ejes de viales.zip') streets <- readOGR('Callejero_ Ejes de viales/call2011.shp', p4s = '+proj=utm +zone=30') streetsMadrid <- streets[streets$CMUN=='079',] streetsMadrid <- spTransform(streetsMadrid, CRS = CRS("+proj=longlat +ellps=WGS84")) library(maptools) ## Lists using the structure accepted by sp.layout, with the polygons, ## lines, and points, and their graphical parameters spDistricts <- list('sp.polygons', distritosMadrid, fill = 'gray97', lwd = 0.3) spStreets <- list('sp.lines', streetsMadrid, lwd = 0.05) spNames <- list(sp.pointLabel, NO2sp, labels = substring(NO2sp$codEst, 7), cex = 0.6, fontfamily = 'Palatino') ## spplot with sp.layout version spplot(NO2sp["classNO2"], col.regions = airPal, cex = dentAQ, edge.col = 'black', alpha = 0.8, ## Boundaries and labels overlaid sp.layout = list(spDistricts, spStreets, spNames), scales = list(draw = TRUE), key.space = NO2key) ## lattice with layer version pNO2 + ## Labels *over* the original figure layer(sp.pointLabel(NO2sp, labels = substring(NO2sp$codEst, 7), cex = 0.8, fontfamily = 'Palatino') ) + ## Polygons and lines *below* (layer_) the figure layer_( { sp.polygons(distritosMadrid, fill = 'gray97', lwd = 0.3) sp.lines(streetsMadrid, lwd = 0.05) }) ################################################################## ## sf and ggplot ################################################################## library(sf) ## Madrid districts distritosMadridSF <- st_read(dsn = 'Distritos de Madrid/', layer = '200001331') distritosMadridSF <- st_transform(distritosMadridSF, crs = "+proj=longlat +ellps=WGS84") ## Madrid streets streetsSF <- st_read(dsn = 'Callejero_ Ejes de viales/', layer = 'call2011', crs = '+proj=longlat +ellps=WGS84') streetsMadridSF <- streetsSF[streetsSF$CMUN=='079',] streetsMadridSF <- st_transform(streetsMadridSF, crs = "+proj=longlat +ellps=WGS84") ggplot()+ ## Layers are drawn sequentially, so the NO2sf layer must be in ## the last place to be on top geom_sf(data = streetsMadridSF, size = 0.05, color = 'lightgray') + geom_sf(data = distritosMadridSF, fill = 'lightgray', alpha = 0.2, size = 0.3, color = 'black') + geom_sf(data = NO2sf, aes(size = classNO2, fill = classNO2), pch = 21, col = 'black') + scale_fill_manual(values = airPal) + scale_size_manual(values = dentAQ * 2) + theme_bw() ################################################################## ## Spatial interpolation ################################################################## library(gstat) ## Sample 10^5 points locations within the bounding box of NO2sp using ## regular sampling airGrid <- spsample(NO2sp, type = 'regular', n = 1e5) ## Convert the SpatialPoints object into a SpatialGrid object gridded(airGrid) <- TRUE ## Compute the IDW interpolation airKrige <- krige(mean ~ 1, NO2sp, airGrid) spplot(airKrige["var1.pred"], ## Variable interpolated col.regions = colorRampPalette(airPal)) + layer({ ## Overlay boundaries and points sp.polygons(distritosMadrid, fill = 'transparent', lwd = 0.3) sp.lines(streetsMadrid, lwd = 0.07) sp.points(NO2sp, pch = 21, alpha = 0.8, fill = 'gray50', col = 'black') }) ################################################################## ## Interactive graphics ################################################################## ################################################################## ## mapView ################################################################## library(mapview) pal <- colorRampPalette(c('springgreen1', 'sienna3', 'gray5'))(100) mapview(NO2sp, zcol = "mean", ## Variable to display cex = "mean", ## Use this variable for the circle sizes col.regions = pal, label = NO2sp$Nombre, legend = TRUE) ################################################################## ## Tooltips with images and graphs ################################################################## img <- paste('images/', NO2sp$codEst, '.jpg', sep = '') mapview(NO2sp, zcol = "mean", cex = "mean", col.regions = pal, label = NO2sp$Nombre, popup = popupImage(img, src = "local"), map.type = "Esri.WorldImagery", legend = TRUE) ## Read the time series airQuality <- read.csv2('data/airQuality.csv') ## We need only NO2 data (codParam 8) NO2 <- subset(airQuality, codParam == 8) ## Time index in a new column NO2$tt <- with(NO2, as.Date(paste(year, month, day, sep = '-'))) ## Stations code stations <- unique(NO2$codEst) ## Loop to create a scatterplot for each station. pList <- lapply(stations, function(i) xyplot(dat ~ tt, data = NO2, subset = (codEst == i), type = 'l', xlab = '', ylab = '') ) mapview(NO2sp, zcol = "mean", cex = "mean", col.regions = pal, label = NO2sp$Nombre, popup = popupGraph(pList), map.type = "Esri.WorldImagery", legend = TRUE) ################################################################## ## Synchronise multiple graphics ################################################################## ## Map of the average value mapMean <- mapview(NO2sp, zcol = "mean", cex = "mean", col.regions = pal, legend = TRUE, map.types = "OpenStreetMap.Mapnik", label = NO2sp$Nombre) ## Map of the median mapMedian <- mapview(NO2sp, zcol = "median", cex = "median", col.regions = pal, legend = TRUE, map.type = "Stamen.Watercolor", label = NO2sp$Nombre) ## Map of the standard deviation mapSD <- mapview(NO2sp, zcol = "sd", cex = "sd", col.regions = pal, legend = TRUE, map.type = "Esri.WorldImagery", label = NO2sp$Nombre) ## All together sync(mapMean, mapMedian, mapSD, ncol = 3) ################################################################## ## GeoJSON and OpenStreepMap ################################################################## library(rgdal) writeOGR(NO2sp, 'data/NO2.geojson', 'NO2sp', driver = 'GeoJSON') ################################################################## ## Keyhole Markup Language ################################################################## library(rgdal) writeOGR(NO2sp, dsn = 'NO2_mean.kml', layer = 'mean', driver = 'KML') library(plotKML) plotKML(NO2sp["mean"], points_names = NO2sp$codEst) ################################################################## ## 3D visualization ################################################################## library(rgl) ## rgl does not understand Spatial* objects NO2df <- as.data.frame(NO2sp) ## Color of each point according to its class airPal <- colorRampPalette(c('springgreen1', 'sienna3', 'gray5'))(5) colorClasses <- airPal[NO2df$classNO2] plot3d(x = NO2df$coords.x1, y = NO2df$coords.x2, z = NO2df$alt, xlab = 'Longitude', ylab = 'Latitude', zlab = 'Altitude', type = 's', col = colorClasses, radius = NO2df$mean/10)
Vector fields
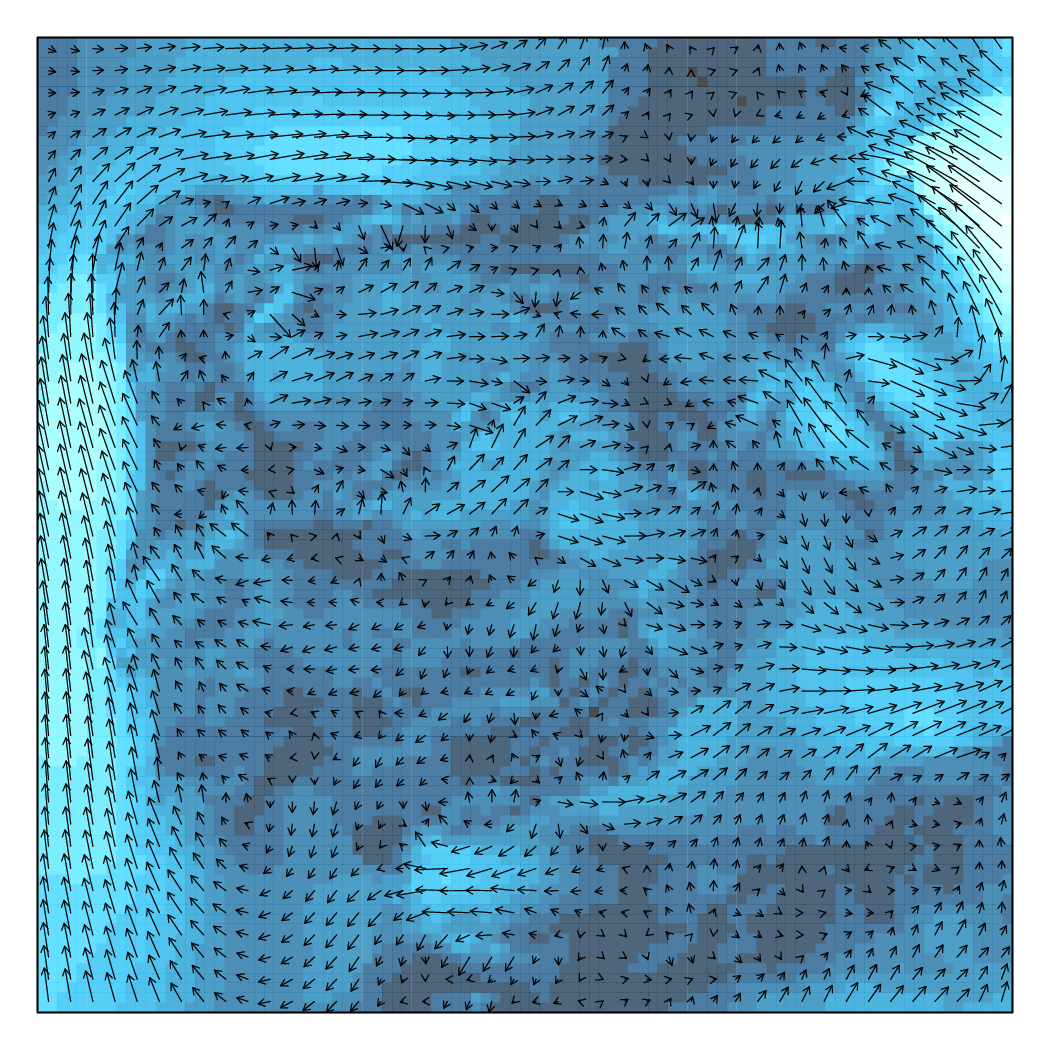
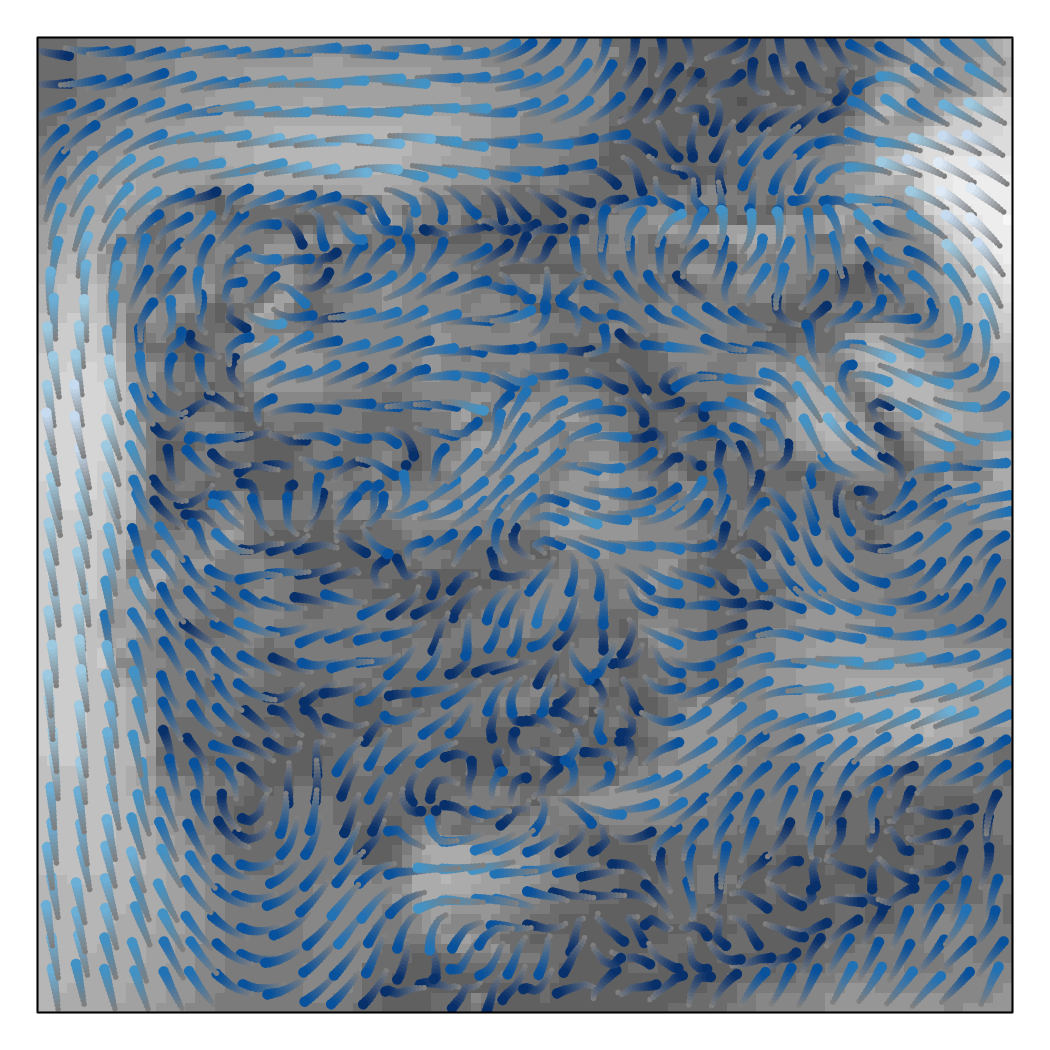
################################################################## ## Initial configuration ################################################################## ## Clone or download the repository and set the working directory ## with setwd to the folder where the repository is located. library(raster) library(rasterVis) wDir <- raster('data/wDir')/180*pi wSpeed <- raster('data/wSpeed') windField <- stack(wSpeed, wDir) names(windField) <- c('magnitude', 'direction') ################################################################## ## Arrow plot ################################################################## vectorTheme <- BTCTheme(regions = list(alpha = 0.7)) vectorplot(windField, isField = TRUE, aspX = 5, aspY = 5, scaleSlope = FALSE, par.settings = vectorTheme, colorkey = FALSE, scales = list(draw = FALSE)) ################################################################## ## Streamlines ################################################################## myTheme <- streamTheme( region = rev(brewer.pal(n = 4, "Greys")), symbol = rev(brewer.pal(n = 9, "Blues"))) streamplot(windField, isField = TRUE, par.settings = myTheme, droplet = list(pc = 12), streamlet = list(L = 5, h = 5), scales = list(draw = FALSE), panel = panel.levelplot.raster)
Physical maps
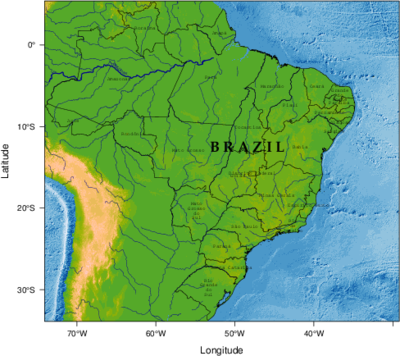
################################################################## ## Initial configuration ################################################################## ## Clone or download the repository and set the working directory ## with setwd to the folder where the repository is located. ################################################################## ## Physical maps ################################################################## library(raster) library(rasterVis) library(rgdal) library(rgeos) library(latticeExtra) library(colorspace) ################################################################## ## Retrieving data from DIVA-GIS, GADM and Natural Earth Data ################################################################## old <- setwd(tempdir()) download.file('http://biogeo.ucdavis.edu/data/gadm2.8/shp/BRA_adm_shp.zip', 'BRA_adm.zip') unzip('BRA_adm.zip') brazilAdm <- readOGR(dsn = '.', layer = 'BRA_adm1') Encoding(levels(brazilAdm$NAME_1)) <- 'latin1' download.file('http://biogeo.ucdavis.edu/data/diva/alt/BRA_alt.zip', 'BRA_alt.zip') unzip('BRA_alt.zip') brazilDEM <- raster('BRA_alt') ## World Water lines (Natural Earth) download.file('http://www.naturalearthdata.com/http//www.naturalearthdata.com/download/10m/physical/ne_10m_rivers_lake_centerlines.zip', 'neRivers.zip') unzip('neRivers.zip') worldlRiv <- readOGR(dsn = '.', layer = 'ne_10m_rivers_lake_centerlines') download.file('http://www.naturalearthdata.com/http//www.naturalearthdata.com/download/10m/raster/OB_LR.zip', 'neSea.zip') unzip('neSea.zip') worldSea <- raster('OB_LR.tif') brazilSea <- crop(worldSea, brazilDEM) setwd(old) ################################################################## ## Intersection of shapefiles and elevation model ################################################################## ## only those features labeled as "River" are needed worldRiv<- worldRiv[worldRiv$featurecla=='River',] ## Define the extent of Brazil as a SpatialPolygons extBrazil <- as(extent(brazilDEM), 'SpatialPolygons') proj4string(extBrazil) <- proj4string(worldRiv) ## and intersect it with worldRiv to extract brazilian rivers ## from the world database brazilRiv <- gIntersection(worldRiv, extBrazil, byid=TRUE) ## and especially the famous Amazonas River amazonas <- worldRiv[worldRiv$name=='Amazonas',] ################################################################## ## Labels ################################################################## ## Locations of labels of each polygon centroids <- coordinates(brazilAdm) ## Location of the "Brazil" label (average of the set of polygons centroids) xyBrazil <- apply(centroids, 2, mean) admNames <- strsplit(as.character(brazilAdm$NAME_1), ' ') admNames <- sapply(admNames, FUN=function(s){ sep=if (length(s)>2) '\n' else ' ' paste(s, collapse=sep) }) ################################################################## ## Overlaying layers of information ################################################################## blueTheme <- rasterTheme(region=brewer.pal(n=9, 'Blues')) seaPlot <- levelplot(brazilSea, par.settings=blueTheme, maxpixels=1e6, panel=panel.levelplot.raster, margin=FALSE, colorkey=FALSE) terrainTheme <- rasterTheme(region=terrain_hcl(15)) altPlot <- levelplot(brazilDEM, par.settings=terrainTheme, maxpixels=1e6, panel=panel.levelplot.raster, margin=FALSE, colorkey=FALSE) amazonasLab <- label(amazonas, 'Amazonas') seaPlot + altPlot + layer({ ## Rivers sp.lines(brazilRiv, col='darkblue', lwd=0.2) ## Amazonas sp.lineLabel(amazonas, amazonasLab, lwd=1, col='darkblue', col.line='darkblue', cex=0.5, fontfamily='Palatino') ## Administrative boundaries sp.polygons(brazilAdm, col='black', lwd=0.2) ## Centroids of administrative boundaries ... panel.points(centroids, col='black') ## ... with their labels panel.pointLabel(centroids, labels=admNames, cex=0.7, fontfamily='Palatino', lineheight=.8) ## Country name panel.text(xyBrazil[1], xyBrazil[2], labels='B R A Z I L', cex=1.5, fontfamily = 'Palatino', fontface=2) }) print(seaPlot, split=c(1, 1, 2, 1), more=TRUE) print(altPlot, split=c(2, 1, 2, 1))
Data
################################################################## ## Initial configuration ################################################################## ## Clone or download the repository and set the working directory ## with setwd to the folder where the repository is located. ################################################################## ## Air Quality in Madrid ################################################################## ## codeStations.csv is extracted from the document ## http://www.mambiente.munimadrid.es/opencms/export/sites/default/calaire/Anexos/INTPHORA-DIA.pdf, ## table of page 3. codEstaciones <- read.csv2('data/codeStations.csv') codURL <- as.numeric(substr(codEstaciones$Codigo, 7, 8)) ## The information of each measuring station is available at its own webpage, defined by codURL URLs <- paste('http://www.mambiente.munimadrid.es/opencms/opencms/calaire/contenidos/estaciones/estacion', codURL, '.html', sep = '') ################################################################## ## Data arrangement ################################################################## library(XML) library(sp) ## Access each webpage, retrieve tables and extract long/lat data coords <- lapply(URLs, function(est){ tables <- readHTMLTable(est) location <- tables[[2]] ## Clean the table content and convert to dms format ub2dms <- function(x){ ch <- as.character(x) ch <- sub(',', '.', ch) ch <- sub('O', 'W', ch) ## Some stations use "O" instead of "W" as.numeric(char2dms(ch, "º", "'", "'' ")) } long <- ub2dms(location[2,1]) lat <- ub2dms(location[2,2]) alt <- as.numeric(sub(' m.', '', location[2, 3])) coords <- data.frame(long = long, lat = lat, alt = alt) coords }) airStations <- cbind(codEstaciones, do.call(rbind, coords)) ## The longitude of "El Pardo" station is wrong (positive instead of negative) airStations$long[22] <- -airStations$long[22] write.csv2(airStations, file = 'data/airStations.csv') rawData <- readLines('data/Datos11.txt') ## This loop reads each line and extracts fields as defined by the ## INTPHORA file: ## http://www.mambiente.munimadrid.es/opencms/export/sites/default/calaire/Anexos/INTPHORA-DIA.pdf datos11 <- lapply(rawData, function(x){ codEst <- substr(x, 1, 8) codParam <- substr(x, 9, 10) codTec <- substr(x, 11, 12) codPeriod <- substr(x, 13, 14) month <- substr(x, 17, 18) dat <- substr(x, 19, nchar(x)) ## "N" used for impossible days (31st April) idxN <- gregexpr('N', dat)[[1]] if (idxN==-1) idxN <- numeric(0) nZeroDays <- length(idxN) day <- seq(1, 31-nZeroDays) ## Substitute V and N with ";" to split data from different days dat <- gsub('[VN]+', ';', dat) dat <- as.numeric(strsplit(dat, ';')[[1]]) ## Only data from valid days dat <- dat[day] res <- data.frame(codEst, codParam, ##codTec, codPeriod, month, day, year = 2016, dat) }) datos11 <- do.call(rbind, datos11) write.csv2(datos11, 'data/airQuality.csv') ################################################################## ## Combine data and spatial locations ################################################################## library(sp) ## Spatial location of stations airStations <- read.csv2('data/airStations.csv') coordinates(airStations) <- ~ long + lat ## Geographical projection proj4string(airStations) <- CRS("+proj=longlat +ellps=WGS84 +datum=WGS84") ## Measurements data airQuality <- read.csv2('data/airQuality.csv') ## Only interested in NO2 NO2 <- airQuality[airQuality$codParam==8, ] NO2agg <- aggregate(dat ~ codEst, data = NO2, FUN = function(x) { c(mean = signif(mean(x), 3), median = median(x), sd = signif(sd(x), 3)) }) NO2agg <- do.call(cbind, NO2agg) NO2agg <- as.data.frame(NO2agg) library(rgdal) library(maptools) ## Link aggregated data with stations to obtain a SpatialPointsDataFrame. ## Codigo and codEst are the stations codes idxNO2 <- match(airStations$Codigo, NO2agg$codEst) NO2sp <- spCbind(airStations[, c('Nombre', 'alt')], NO2agg[idxNO2, ]) ## Save the result writeOGR(NO2sp, dsn = 'data/', layer = 'NO2sp', driver = 'ESRI Shapefile') ################################################################## ## Photographs of the stations ################################################################## library(XML) old <- setwd('images') for (i in 1:nrow(NO2df)) { codEst <- NO2df[i, "codEst"] ## Webpage of each station codURL <- as.numeric(substr(codEst, 7, 8)) rootURL <- 'http://www.mambiente.munimadrid.es' stationURL <- paste(rootURL, '/opencms/opencms/calaire/contenidos/estaciones/estacion', codURL, '.html', sep = '') content <- htmlParse(stationURL, encoding = 'utf8') ## Extracted with http://www.selectorgadget.com/ xPath <- '//*[contains(concat( " ", @class, " " ), concat( " ", "imagen_1", " " ))]' imageStation <- getNodeSet(content, xPath)[[1]] imageURL <- xmlAttrs(imageStation)[1] imageURL <- paste(rootURL, imageURL, sep = '') download.file(imageURL, destfile = paste(codEst, '.jpg', sep = '')) } setwd(old) ################################################################## ## Spanish General Elections ################################################################## dat2016 <- read.csv('data/GeneralSpanishElections2016.csv') census <- dat2016$Total.censo.electoral validVotes <- dat2016$Votos.válidos ## Election results per political party and municipality votesData <- dat2016[, -(1:13)] ## Abstention as an additional party votesData$ABS <- census - validVotes ## UP is a coalition of several parties UPcols <- grep("PODEMOS|ECP", names(votesData)) votesData$UP <- rowSums(votesData[, UPcols]) votesData[, UPcols] <- NULL ## Winner party at each municipality whichMax <- apply(votesData, 1, function(x)names(votesData)[which.max(x)]) ## Results of the winner party at each municipality Max <- apply(votesData, 1, max) ## OTH for everything but PP, PSOE, UP, Cs, and ABS whichMax[!(whichMax %in% c('PP', 'PSOE', 'UP', 'C.s', 'ABS'))] <- 'OTH' ## Percentage of votes with the electoral census pcMax <- Max/census * 100 ## Province-Municipality code. sprintf formats a number with leading zeros. PROVMUN <- with(dat2016, paste(sprintf('%02d', Código.de.Provincia), sprintf('%03d', Código.de.Municipio), sep="")) votes2016 <- data.frame(PROVMUN, whichMax, Max, pcMax) write.csv(votes2016, 'data/votes2016.csv', row.names = FALSE) ################################################################## ## Administrative boundaries ################################################################## library(sp) library(rgdal) old <- setwd(tempdir()) download.file('ftp://www.ine.es/pcaxis/mapas_completo_municipal.rar', 'mapas_completo_municipal.rar') system2('unrar', c('e', 'mapas_completo_municipal.rar')) spMap <- readOGR("esp_muni_0109.shp", p4s = "+proj=utm +zone=30 +ellps=GRS80 +units=m +no_defs") Encoding(levels(spMap$NOMBRE)) <- "latin1" setwd(old) ## dissolve repeated polygons spPols <- unionSpatialPolygons(spMap, spMap$PROVMUN) votes2016 <- read.csv('data/votes2016.csv', colClasses = c('factor', 'factor', 'numeric', 'numeric')) ## Match polygons and data using ID slot and PROVMUN column IDs <- sapply(spPols@polygons, function(x)x@ID) idx <- match(IDs, votes2016$PROVMUN) ##Places without information idxNA <- which(is.na(idx)) ##Information to be added to the SpatialPolygons object dat2add <- votes2016[idx, ] ## SpatialPolygonsDataFrame uses row names to match polygons with data row.names(dat2add) <- IDs spMapVotes <- SpatialPolygonsDataFrame(spPols, dat2add) ## Drop those places without information spMapVotes0 <- spMapVotes[-idxNA, ] ## Save the result writeOGR(spMapVotes0, dsn = 'data/', layer = 'spMapVotes0', drive = 'ESRI Shapefile') ## Extract Canarias islands from the SpatialPolygons object canarias <- sapply(spMapVotes0@polygons, function(x)substr(x@ID, 1, 2) %in% c("35", "38")) peninsula <- spMapVotes0[!canarias,] island <- spMapVotes0[canarias,] ## Shift the island extent box to position them at the bottom right corner dy <- bbox(peninsula)[2,1] - bbox(island)[2,1] dx <- bbox(peninsula)[1,2] - bbox(island)[1,2] island2 <- elide(island, shift = c(dx, dy)) bbIslands <- bbox(island2) proj4string(island2) <- proj4string(peninsula) ## Bind Peninsula (without islands) with shifted islands spMapVotes <- rbind(peninsula, island2) ## Save the result writeOGR(spMapVotes, dsn = 'data/', layer = 'spMapVotes', drive = 'ESRI Shapefile') ################################################################## ## CM SAF ################################################################## library(raster) tmp <- tempdir() unzip('data/SISmm2008_CMSAF.zip', exdir = tmp) filesCMSAF <- dir(tmp, pattern = 'SISmm') SISmm <- stack(paste(tmp, filesCMSAF, sep = '/')) ## CM-SAF data is average daily irradiance (W/m2). Multiply by 24 ## hours to obtain daily irradiation (Wh/m2) SISmm <- SISmm * 24 ## Monthly irradiation: each month by the corresponding number of days daysMonth <- c(31, 29, 31, 30, 31, 30, 31, 31, 30, 31, 30, 31) SISm <- SISmm * daysMonth / 1000 ## kWh/m2 ## Annual average SISav <- sum(SISm)/sum(daysMonth) writeRaster(SISav, file = 'SISav') library(raster) ## http://neo.sci.gsfc.nasa.gov/Search.html?group=64 pop <- raster('875430rgb-167772161.0.FLOAT.TIFF') ## http://neo.sci.gsfc.nasa.gov/Search.html?group=20 landClass <- raster('241243rgb-167772161.0.TIFF')