Time Series - Displaying time series, spatial and space-time data with R
Table of Contents
Time on the horizontal axis
Time as a conditioning or grouping variable
Time as a complementary variable
Code
Time on the horizontal axis
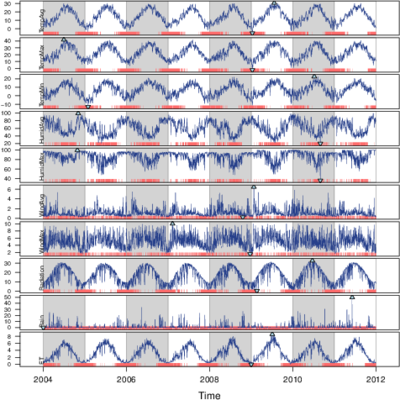
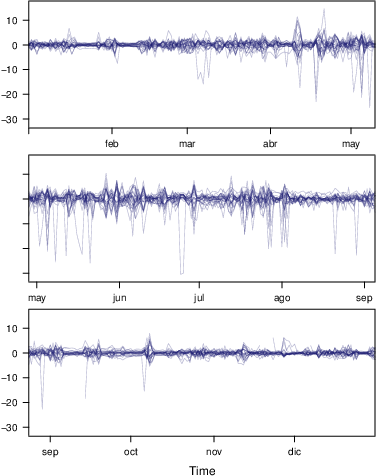
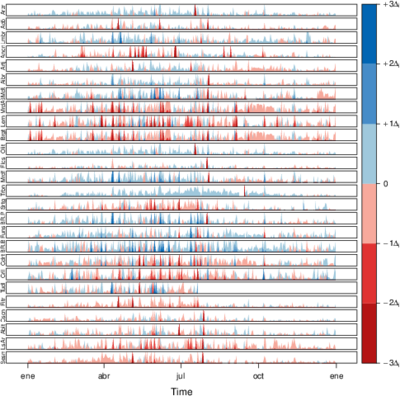
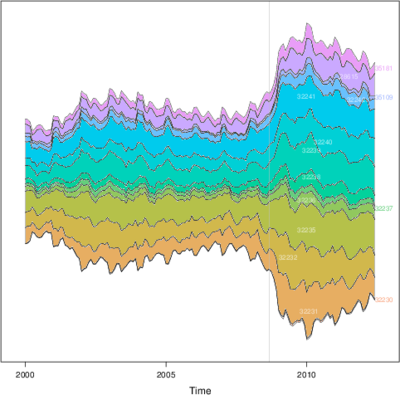
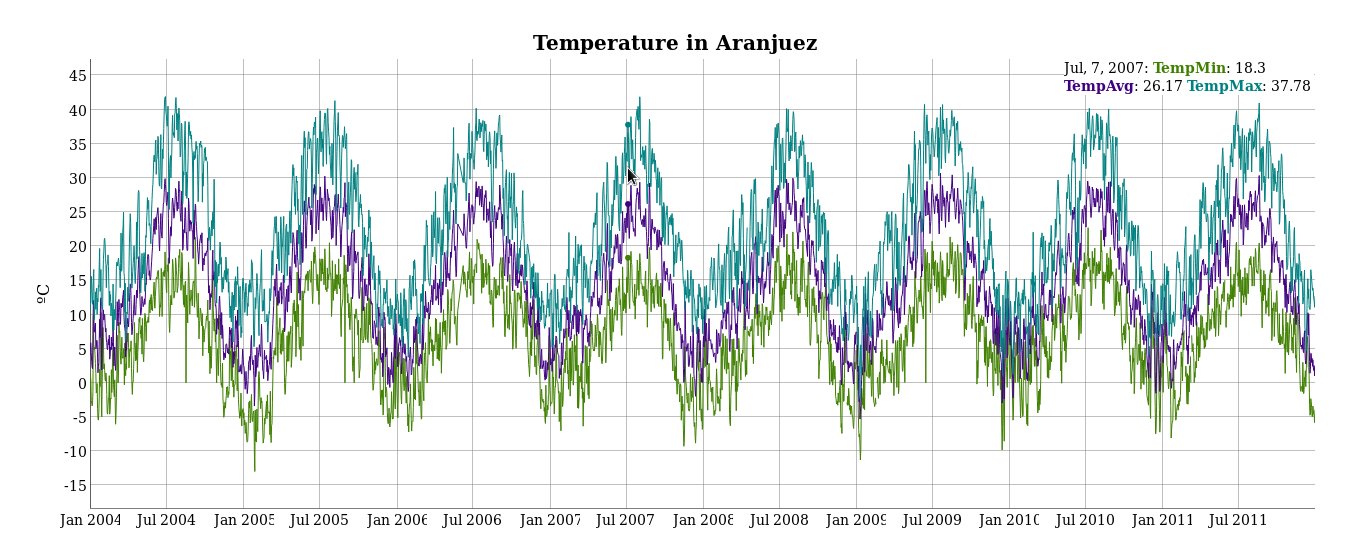
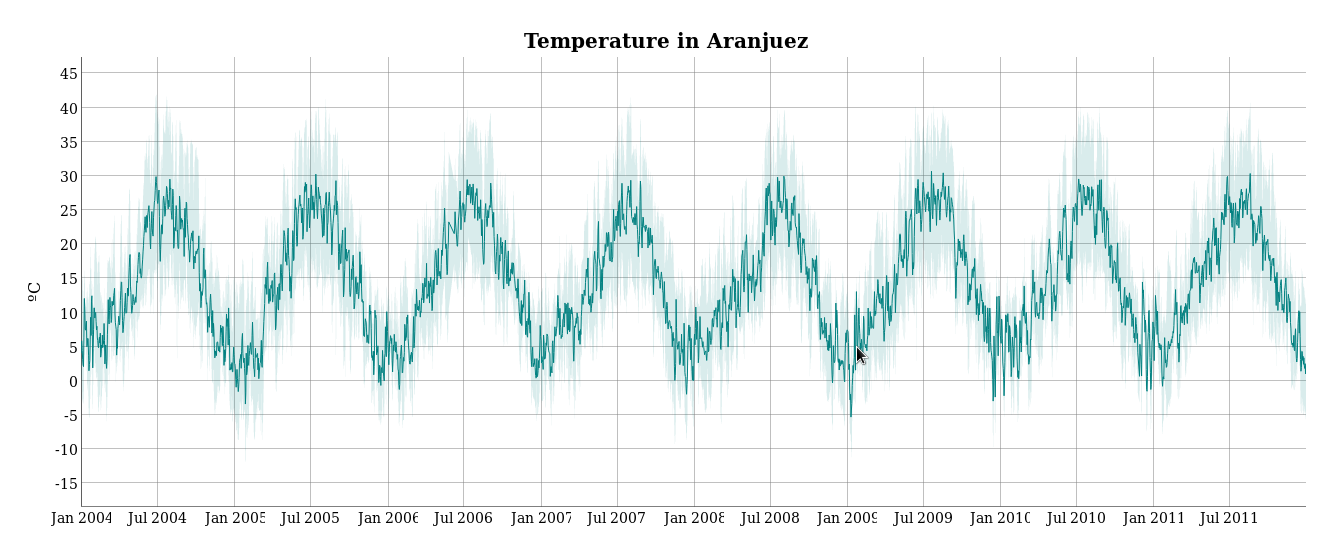
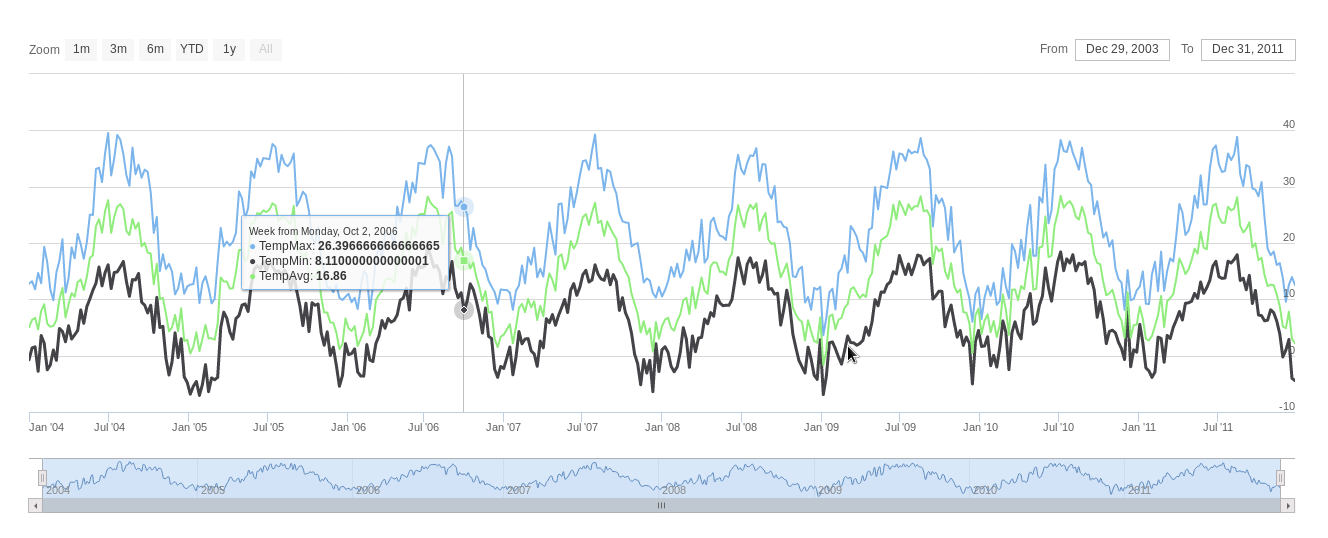
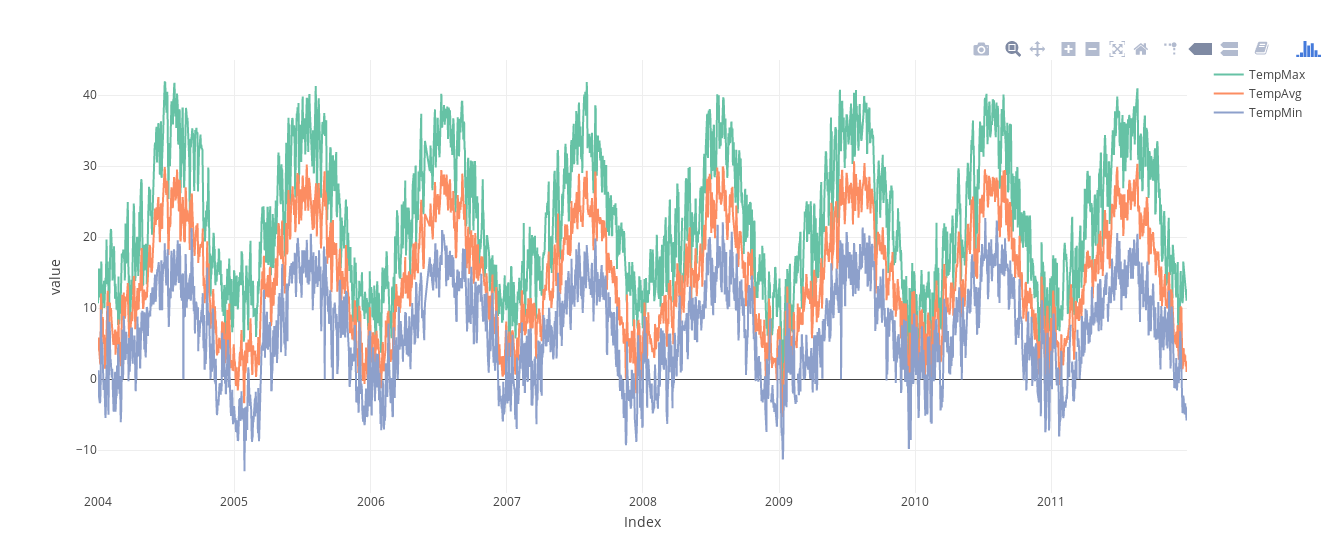
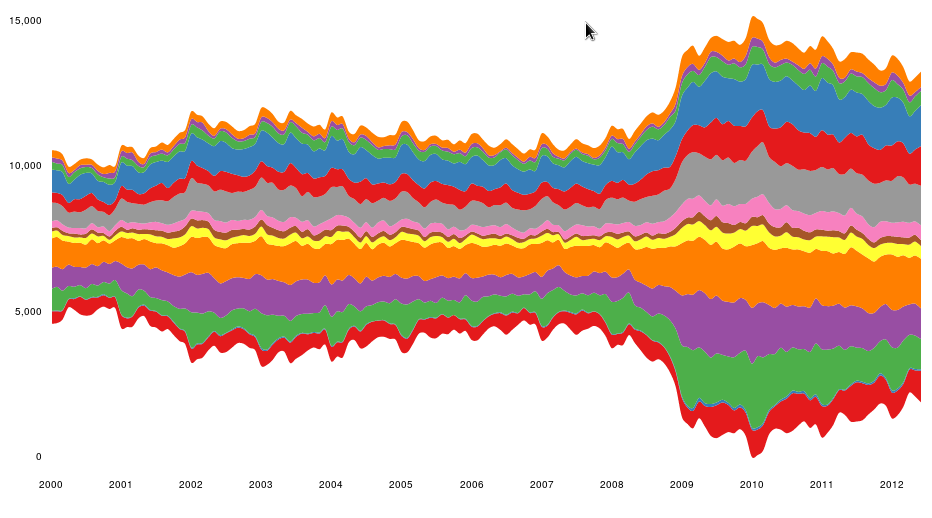
################################################################## ## Initial configuration ################################################################## ## Clone or download the repository and set the working directory ## with setwd to the folder where the repository is located. library(lattice) library(ggplot2) ## latticeExtra must be loaded after ggplot2 to prevent masking of its ## `layer` function. library(latticeExtra) source('configLattice.R') ################################################################## ################################################################## ## Time graph of variables with different scales ################################################################## library(zoo) load('data/aranjuez.RData') ## The layout argument arranges panels in rows xyplot(aranjuez, layout = c(1, ncol(aranjuez))) autoplot(aranjuez) + facet_free() ################################################################## ## Annotations to enhance the time graph ################################################################## ## lattice version ## Auxiliary function to extract the year value of a POSIXct time ## index Year <- function(x)format(x, "%Y") xyplot(aranjuez, layout = c(1, ncol(aranjuez)), strip = FALSE, scales = list(y = list(cex = 0.6, rot = 0)), panel = function(x, y, ...){ ## Alternation of years panel.xblocks(x, Year, col = c("lightgray", "white"), border = "darkgray") ## Values under the average highlighted with red regions panel.xblocks(x, y < mean(y, na.rm = TRUE), col = "indianred1", height = unit(0.1, 'npc')) ## Time series panel.lines(x, y, col = 'royalblue4', lwd = 0.5, ...) ## Label of each time series panel.text(x[1], min(y, na.rm = TRUE), names(aranjuez)[panel.number()], cex = 0.6, adj = c(0, 0), srt = 90, ...) ## Triangles to point the maxima and minima idxMax <- which.max(y) panel.points(x[idxMax], y[idxMax], col = 'black', fill = 'lightblue', pch = 24) idxMin <- which.min(y) panel.points(x[idxMin], y[idxMin], col = 'black', fill = 'lightblue', pch = 25) }) ## ggplot2 version timeIdx <- index(aranjuez) aranjuezLong <- fortify(aranjuez, melt = TRUE) summary(aranjuezLong) ## Values below mean are negative after being centered scaled <- fortify(scale(aranjuez, scale = FALSE), melt = TRUE) ## The 'scaled' column is the result of the centering. ## The new 'Value' column store the original values. scaled <- transform(scaled, scaled = Value, Value = aranjuezLong$Value) underIdx <- which(scaled$scaled <= 0) ## 'under' is the subset of values below the average under <- scaled[underIdx,] library(xts) ep <- endpoints(timeIdx, on = 'years') ep <- ep[-1] N <- length(ep) ## 'tsp' is start and 'tep' is the end of each band. One of each two ## years are selected. tep <- timeIdx[ep[seq(1, N, 2)] + 1] tsp <- timeIdx[ep[seq(2, N, 2)]] minIdx <- timeIdx[apply(aranjuez, 2, which.min)] minVals <- apply(aranjuez, 2, min, na.rm = TRUE) mins <- data.frame(Index = minIdx, Value = minVals, Series = names(aranjuez)) maxIdx <- timeIdx[apply(aranjuez, 2, which.max)] maxVals <- apply(aranjuez, 2, max, na.rm = TRUE) maxs <- data.frame(Index = maxIdx, Value = maxVals, Series = names(aranjuez)) ggplot(data = aranjuezLong, aes(Index, Value)) + ## Time series of each variable geom_line(colour = "royalblue4", lwd = 0.5) + ## Year bands annotate('rect', xmin = tsp, xmax = tep, ymin = -Inf, ymax = Inf, alpha = 0.4) + ## Values below average geom_rug(data = under, sides = 'b', col = 'indianred1') + ## Minima geom_point(data = mins, pch = 25) + ## Maxima geom_point(data = maxs, pch = 24) + ## Axis labels and theme definition labs(x = 'Time', y = NULL) + theme_bw() + ## Each series is displayed in a different panel with an ## independent y scale facet_free() ################################################################## ## Time series of variables with the same scale ################################################################## load('data/navarra.RData') avRad <- zoo(rowMeans(navarra, na.rm = 1), index(navarra)) pNavarra <- xyplot(navarra - avRad, superpose = TRUE, auto.key = FALSE, lwd = 0.5, alpha = 0.3, col = 'midnightblue') pNavarra ################################################################## ## Aspect ratio and rate of change ################################################################## xyplot(navarra - avRad, aspect = 'xy', cut = list(n = 3, overlap = 0.1), strip = FALSE, superpose = TRUE, auto.key = FALSE, lwd = 0.5, alpha = 0.3, col = 'midnightblue') ################################################################## ## The horizon graph ################################################################## library(latticeExtra) horizonplot(navarra - avRad, layout = c(1, ncol(navarra)), origin = 0, ## Deviations in each panel are calculated ## from this value colorkey = TRUE, col.regions = brewer.pal(6, "RdBu")) ################################################################## ## Time graph of the differences between a time series and a reference ################################################################## Ta <- aranjuez$TempAvg timeIndex <- index(aranjuez) longTa <- ave(Ta, format(timeIndex, '%j')) diffTa <- (Ta - longTa) xyplot(cbind(Ta, longTa, diffTa), col = c('darkgray', 'red', 'midnightblue'), superpose = TRUE, auto.key = list(space = 'right'), screens = c(rep('Average Temperature', 2), 'Differences')) years <- unique(format(timeIndex, '%Y')) horizonplot(diffTa, cut = list(n = 8, overlap = 0), colorkey = TRUE, col.regions = brewer.pal(6, "RdBu"), layout = c(1, 8), scales = list(draw = FALSE, y = list(relation = 'same')), origin = 0, strip.left = FALSE) + layer(grid.text(years[panel.number()], x = 0, y = 0.1, gp = gpar(cex = 0.8), just = "left")) year <- function(x)as.numeric(format(x, '%Y')) day <- function(x)as.numeric(format(x, '%d')) month <- function(x)as.numeric(format(x, '%m')) myTheme <- modifyList(custom.theme(region = brewer.pal(9, 'RdBu')), list( strip.background = list(col = 'gray'), panel.background = list(col = 'gray'))) maxZ <- max(abs(diffTa)) levelplot(diffTa ~ day(timeIndex) * year(timeIndex) | factor(month(timeIndex)), at = pretty(c(-maxZ, maxZ), n = 8), colorkey = list(height = 0.3), layout = c(1, 12), strip = FALSE, strip.left = TRUE, xlab = 'Day', ylab = 'Month', par.settings = myTheme) df <- data.frame(Vals = diffTa, Day = day(timeIndex), Year = year(timeIndex), Month = month(timeIndex)) library(scales) ## The packages scales is needed for the pretty_breaks function. ggplot(data = df, aes(fill = Vals, x = Day, y = Year)) + facet_wrap(~ Month, ncol = 1, strip.position = 'left') + scale_y_continuous(breaks = pretty_breaks()) + scale_fill_distiller(palette = 'RdBu', direction = 1) + geom_raster() + theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank()) ################################################################## ## Stacked graphs ################################################################## load('data/unemployUSA.RData') xyplot(unemployUSA, superpose = TRUE, par.settings = custom.theme, auto.key = list(space = 'right')) library(scales) ## scale_x_yearmon needs scales::pretty_breaks autoplot(unemployUSA, facets = NULL, geom = 'area') + geom_area(aes(fill = Series)) + scale_x_yearmon() panel.flow <- function(x, y, groups, origin, ...) { dat <- data.frame(x = x, y = y, groups = groups) (dataframe) nVars <- nlevels(groups) groupLevels <- levels(groups) ## From long to wide yWide <- unstack(dat, y~groups) (yunstack) ## Where are the maxima of each variable located? We will use ## them to position labels. idxMaxes <- apply(yWide, 2, which.max) ##Origin calculated following Havr.eHetzler.ea2002 if (origin=='themeRiver') origin = -1/2*rowSums(yWide) else origin = 0 yWide <- cbind(origin = origin, yWide) ## Cumulative sums to define the polygon yCumSum <- t(apply(yWide, 1, cumsum)) (ycumsum) Y <- as.data.frame(sapply(seq_len(nVars), (yPolygon) function(iCol)c(yCumSum[,iCol+1], rev(yCumSum[,iCol])))) names(Y) <- levels(groups) ## Back to long format, since xyplot works that way y <- stack(Y)$values (yStack) ## Similar but easier for x xWide <- unstack(dat, x~groups) (xunstack) x <- rep(c(xWide[,1], rev(xWide[,1])), nVars) (xPolygon) ## Groups repeated twice (upper and lower limits of the polygon) groups <- rep(groups, each = 2) (groups) ## Graphical parameters superpose.polygon <- trellis.par.get("superpose.polygon") (gpar) col = superpose.polygon$col border = superpose.polygon$border lwd = superpose.polygon$lwd ## Draw polygons for (i in seq_len(nVars)){ (forVars) xi <- x[groups==groupLevels[i]] yi <- y[groups==groupLevels[i]] panel.polygon(xi, yi, border = border, lwd = lwd, col = col[i]) } ## Print labels for (i in seq_len(nVars)){ xi <- x[groups==groupLevels[i]] yi <- y[groups==groupLevels[i]] N <- length(xi)/2 ## Height available for the label h <- unit(yi[idxMaxes[i]], 'native') - unit(yi[idxMaxes[i] + 2*(N-idxMaxes[i]) +1], 'native') ##...converted to "char" units hChar <- convertHeight(h, 'char', TRUE) ## If there is enough space and we are not at the first or ## last variable, then the label is printed inside the polygon. if((hChar >= 1) && !(i %in% c(1, nVars))){ (hChar) grid.text(groupLevels[i], xi[idxMaxes[i]], (yi[idxMaxes[i]] + yi[idxMaxes[i] + 2*(N-idxMaxes[i]) +1])/2, gp = gpar(col = 'white', alpha = 0.7, cex = 0.7), default.units = 'native') } else { ## Elsewhere, the label is printed outside grid.text(groupLevels[i], xi[N], (yi[N] + yi[N+1])/2, gp = gpar(col = col[i], cex = 0.7), just = 'left', default.units = 'native') } } } prepanel.flow <- function(x, y, groups, origin,...) { dat <- data.frame(x = x, y = y, groups = groups) nVars <- nlevels(groups) groupLevels <- levels(groups) yWide <- unstack(dat, y~groups) if (origin=='themeRiver') origin = -1/2*rowSums(yWide) else origin = 0 yWide <- cbind(origin = origin, yWide) yCumSum <- t(apply(yWide, 1, cumsum)) list(xlim = range(x), (xlim) ylim = c(min(yCumSum[,1]), max(yCumSum[,nVars+1])), (ylim) dx = diff(x), (dx) dy = diff(c(yCumSum[,-1]))) } library(colorspace) ## We will use a qualitative palette from colorspace nCols <- ncol(unemployUSA) pal <- rainbow_hcl(nCols, c = 70, l = 75, start = 30, end = 300) myTheme <- custom.theme(fill = pal, lwd = 0.2) sep2008 <- as.numeric(as.yearmon('2008-09')) xyplot(unemployUSA, superpose = TRUE, auto.key = FALSE, panel = panel.flow, prepanel = prepanel.flow, origin = 'themeRiver', scales = list(y = list(draw = FALSE)), par.settings = myTheme) + layer(panel.abline(v = sep2008, col = 'gray', lwd = 0.7)) ################################################################## ## Panel and prepanel functions to implement the ThemeRiver with =xyplot= ################################################################## library(dygraphs) dyTemp <- dygraph(aranjuez[, c("TempMin", "TempAvg", "TempMax")], main = "Temperature in Aranjuez", ylab = "ºC") dyTemp dyTemp %>% dyHighlight(highlightSeriesBackgroundAlpha = 0.2, highlightSeriesOpts = list(strokeWidth = 2)) dygraph(aranjuez[, c("TempMin", "TempAvg", "TempMax")], main = "Temperature in Aranjuez", ylab = "ºC") %>% dySeries(c("TempMin", "TempAvg", "TempMax"), label = "Temperature") library(highcharter) library(xts) aranjuezXTS <- as.xts(aranjuez) highchart(type = "stock") %>% hc_add_series(name = 'TempMax', aranjuezXTS[, "TempMax"]) %>% hc_add_series(name = 'TempMin', aranjuezXTS[, "TempMin"]) %>% hc_add_series(name = 'TempAvg', aranjuezXTS[, "TempAvg"]) aranjuezDF <- fortify(aranjuez[, c("TempMax", "TempAvg", "TempMin")], melt = TRUE) summary(aranjuezDF) library(plotly) plot_ly(aranjuezDF) %>% add_lines(x = ~ Index, y = ~ Value, color = ~ Series) unemployDF <- fortify(unemployUSA, melt = TRUE) head(unemployDF) ## remotes::install_github("hrbrmstr/streamgraph") library(streamgraph) streamgraph(unemployDF, key = "Series", value = "Value", date = "Index") %>% sg_axis_x(1, "year", "%Y") %>% sg_fill_brewer("Set1")
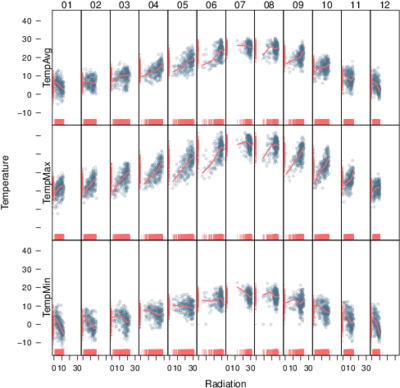
Time as a conditioning or grouping variable
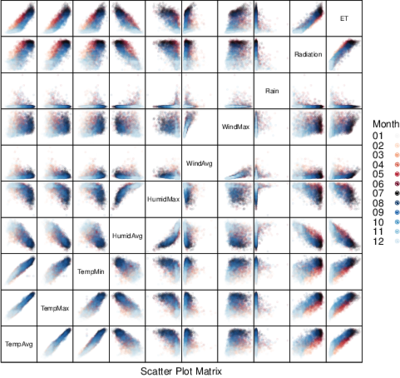
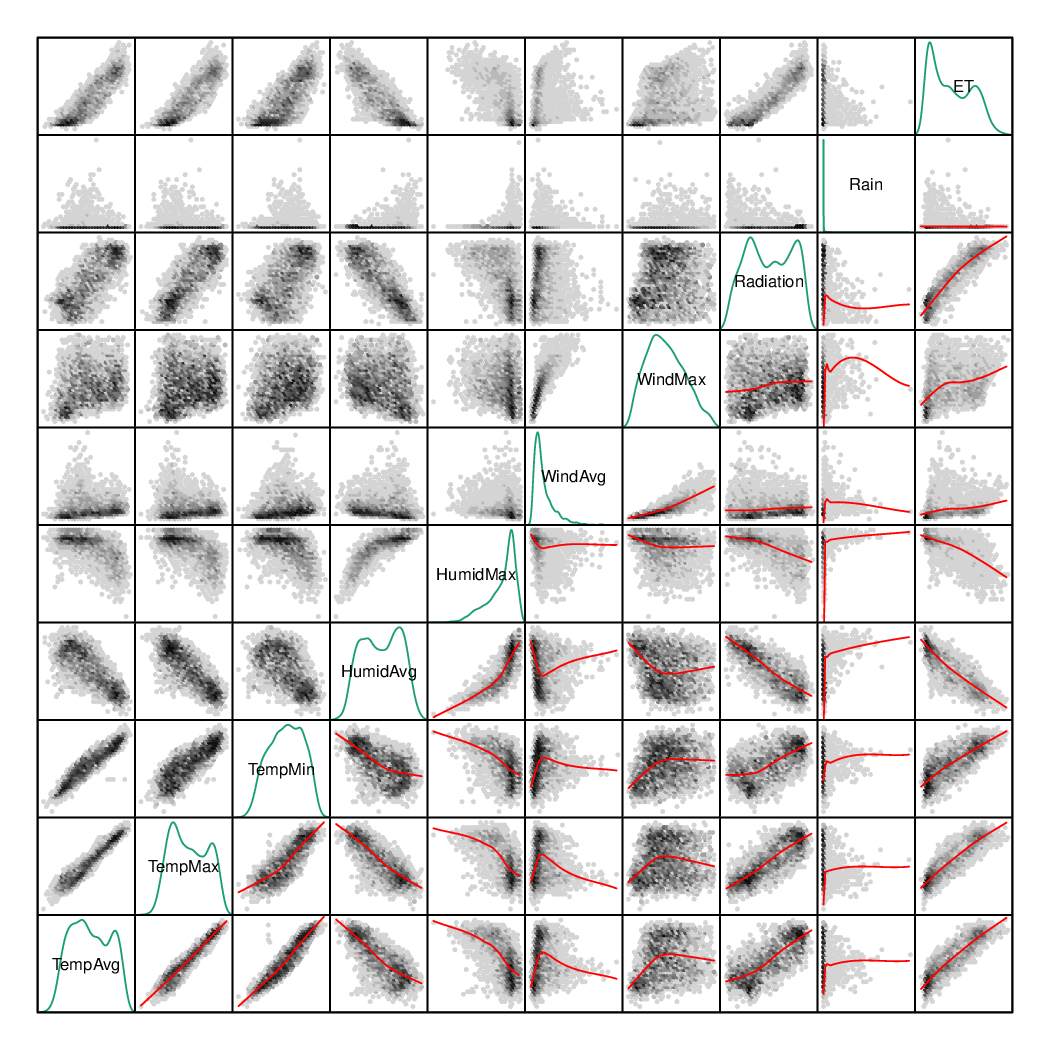
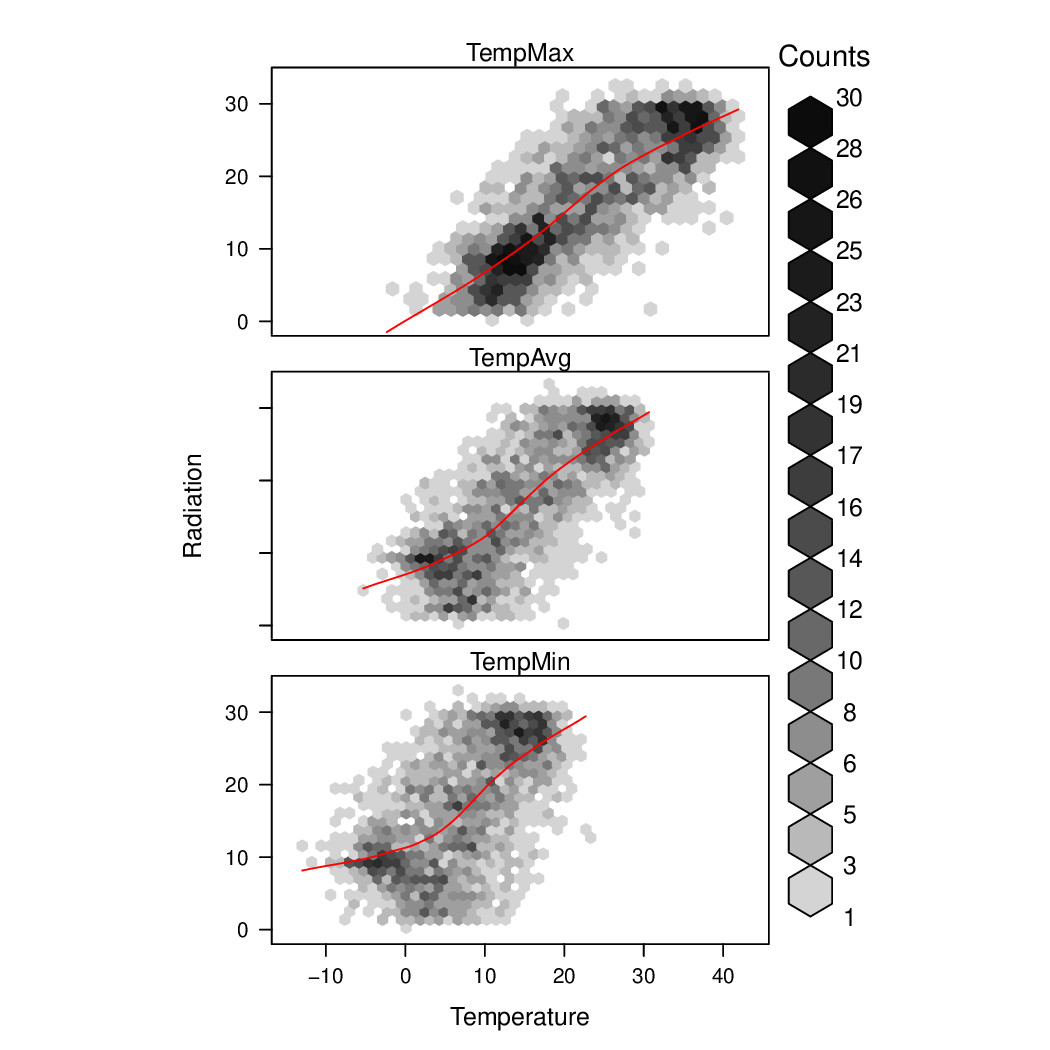
################################################################## ## Initial configuration ################################################################## ## Clone or download the repository and set the working directory ## with setwd to the folder where the repository is located. library(lattice) library(ggplot2) ## latticeExtra must be loaded after ggplot2 to prevent masking of its ## `layer` function. library(latticeExtra) source('configLattice.R') ################################################################## ################################################################## ## Scatterplot matrix: time as a grouping variable ################################################################## library(zoo) load('data/aranjuez.RData') aranjuezDF <- as.data.frame(aranjuez) aranjuezDF$Month <- format(index(aranjuez), '%m') ## Red-Blue palette with black added (12 colors) colors <- c(brewer.pal(n = 11, 'RdBu'), '#000000') ## Rearrange according to months (darkest for summer) colors <- colors[c(6:1, 12:7)] splom(~ aranjuezDF[1:10], groups = aranjuezDF$Month, auto.key = list(space = 'right', title = 'Month', cex.title = 1), pscale = 0, varname.cex = 0.7, xlab = '', par.settings = custom.theme(symbol = colors, pch = 19), cex = 0.3, alpha = 0.1) trellis.focus('panel', 1, 1) idx <- panel.link.splom(pch = 13, cex = 0.6, col = 'green') aranjuez[idx,] library(GGally) ggpairs(aranjuezDF, columns = 1:10, ## Do not include "Month" upper = list(continuous = "points"), mapping = aes(colour = Month, alpha = 0.1)) ################################################################## ## Hexagonal binning ################################################################## library(hexbin) splom(~as.data.frame(aranjuez), panel = panel.hexbinplot, diag.panel = function(x, ...){ yrng <- current.panel.limits()$ylim d <- density(x, na.rm = TRUE) d$y <- with(d, yrng[1] + 0.95 * diff(yrng) * y / max(y)) panel.lines(d) diag.panel.splom(x, ...) }, lower.panel = function(x, y, ...){ panel.hexbinplot(x, y, ...) panel.loess(x, y, ..., col = 'red') }, xlab = '', pscale = 0, varname.cex = 0.7) library(reshape2) aranjuezRshp <- melt(aranjuezDF, measure.vars = c('TempMax', 'TempAvg', 'TempMin'), variable.name = 'Statistic', value.name = 'Temperature') summary(aranjuezRshp) hexbinplot(Radiation ~ Temperature | Statistic, data = aranjuezRshp, layout = c(1, 3)) + layer(panel.loess(..., col = 'red')) ggplot(data = aranjuezRshp, aes(Temperature, Radiation)) + stat_binhex(ncol = 1) + stat_smooth(se = FALSE, method = 'loess', col = 'red') + facet_wrap(~ Statistic, ncol = 1) + theme_bw() ################################################################## ## Scatterplot with time as a conditioning variable ################################################################## ggplot(data = aranjuezRshp, aes(Radiation, Temperature)) + facet_grid(Statistic ~ Month) + geom_point(col = 'skyblue4', pch = 19, cex = 0.5, alpha = 0.3) + geom_rug() + stat_smooth(se = FALSE, method = 'loess', col = 'indianred1', lwd = 1.2) + theme_bw() useOuterStrips( xyplot(Temperature ~ Radiation | Month * Statistic, data = aranjuezRshp, between = list(x = 0), col = 'skyblue4', pch = 19, cex = 0.5, alpha = 0.3)) + layer({ panel.rug(..., col.line = 'indianred1', end = 0.05, alpha = 0.6) panel.loess(..., col = 'indianred1', lwd = 1.5, alpha = 1) })
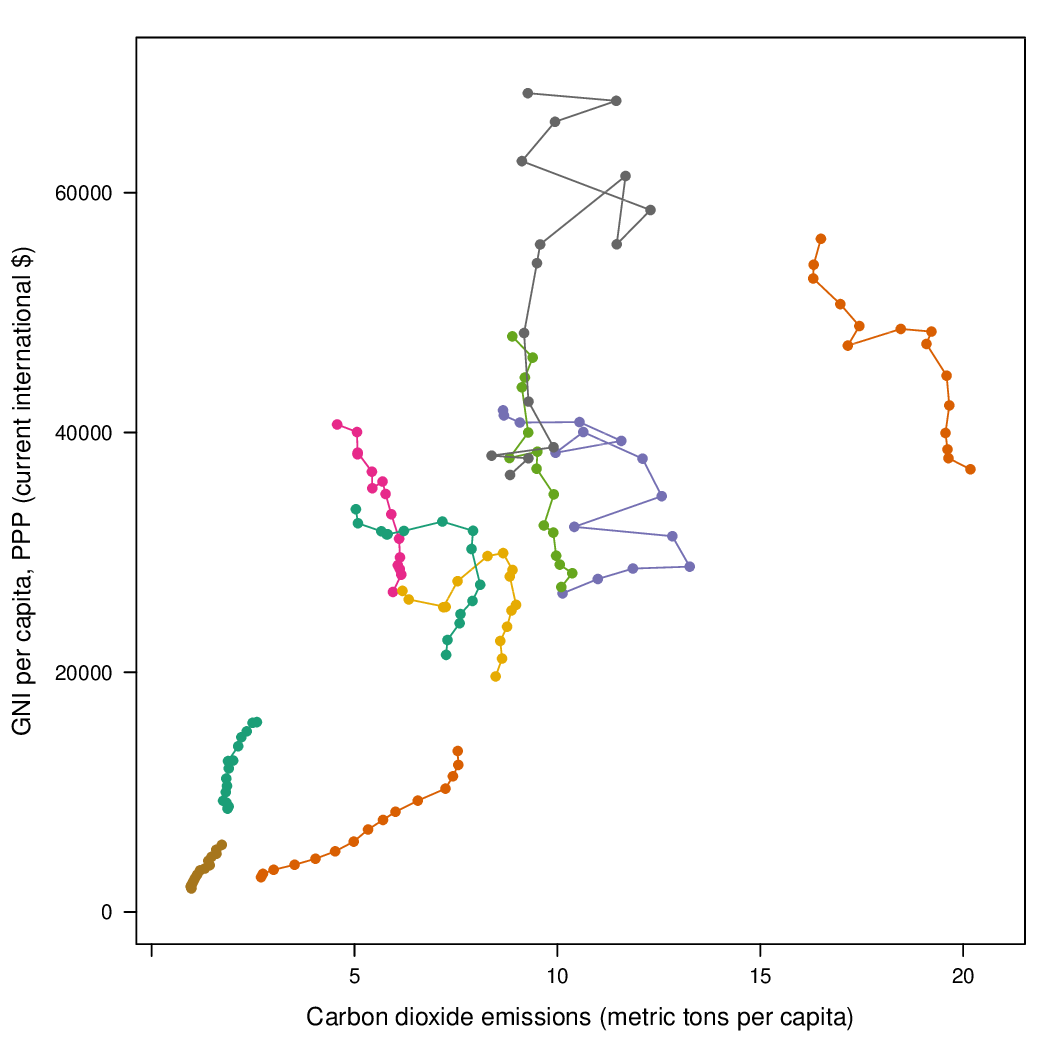
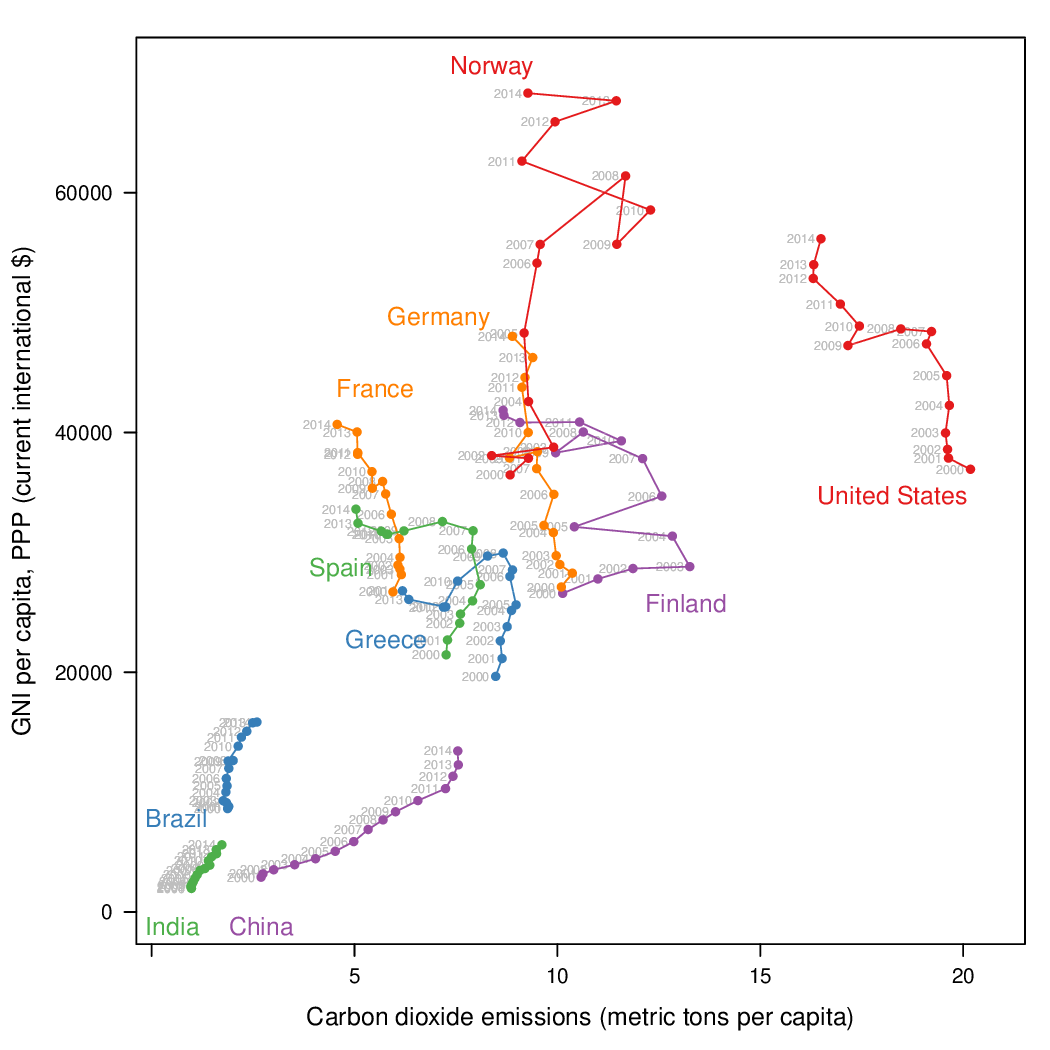
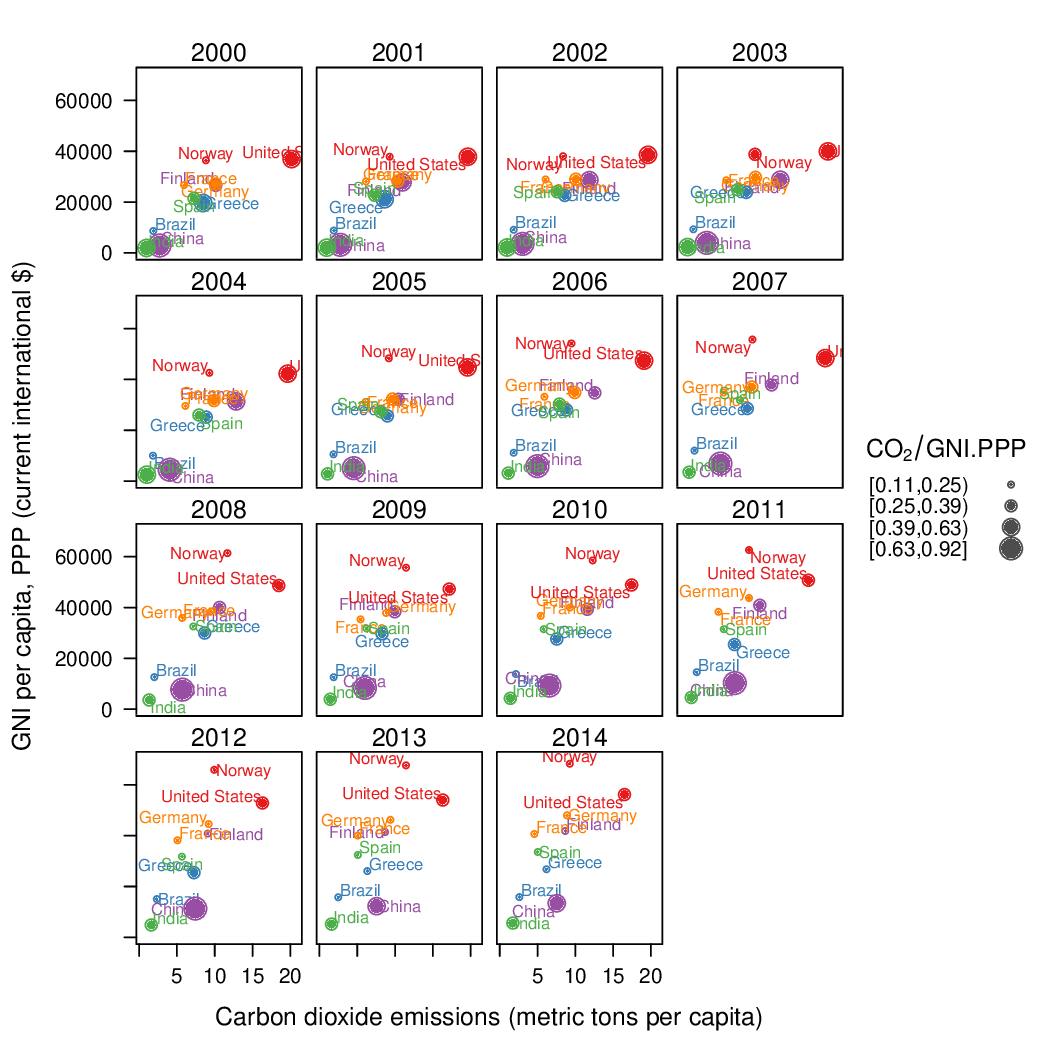
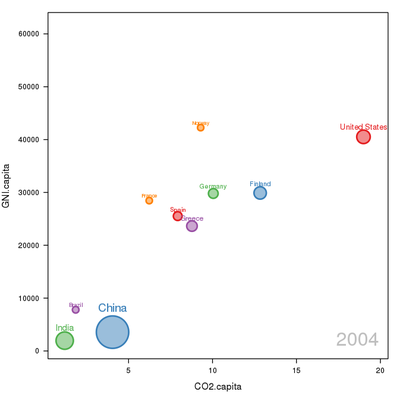
Time as a complementary variable
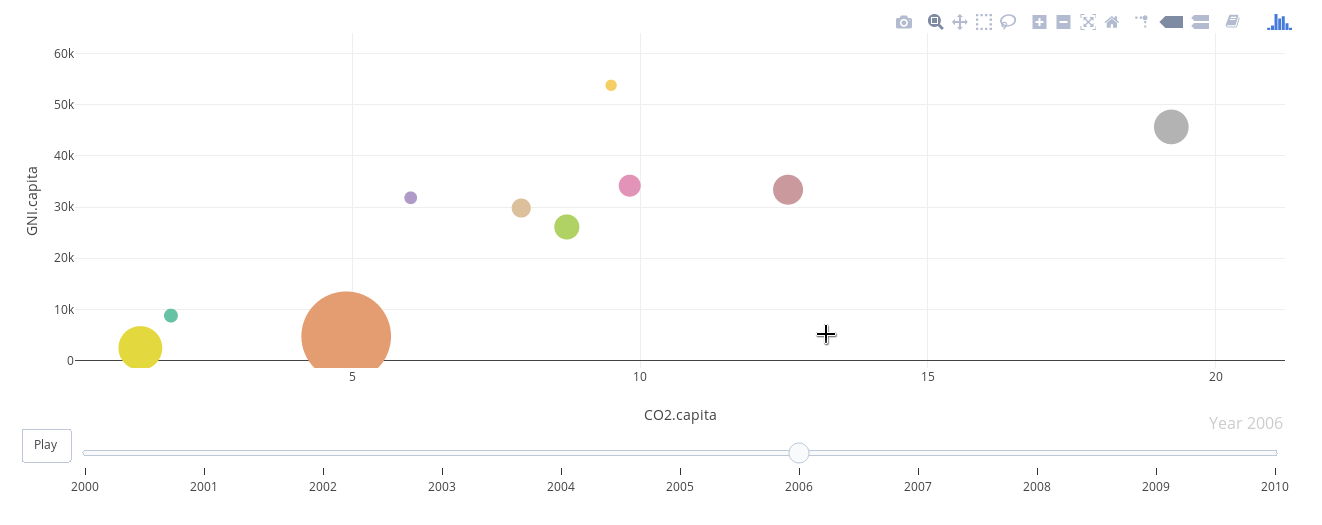
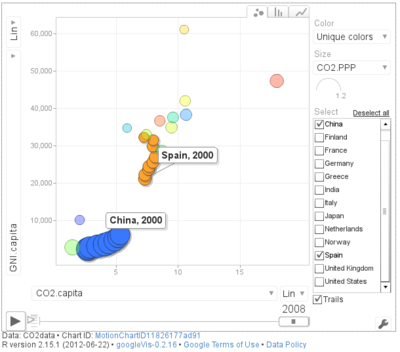
################################################################## ## Initial configuration ################################################################## ## Clone or download the repository and set the working directory ## with setwd to the folder where the repository is located. library(lattice) library(ggplot2) ## latticeExtra must be loaded after ggplot2 to prevent masking of its ## `layer` function. library(latticeExtra) source('configLattice.R') ################################################################## ################################################################## ## Polylines ################################################################## library(zoo) load('data/CO2.RData') ## lattice version xyplot(GNI.capita ~ CO2.capita, data = CO2data, xlab = "Carbon dioxide emissions (metric tons per capita)", ylab = "GNI per capita, PPP (current international $)", groups = Country.Name, type = 'b') ## ggplot2 version ggplot(data = CO2data, aes(x = CO2.capita, y = GNI.capita, color = Country.Name)) + xlab("Carbon dioxide emissions (metric tons per capita)") + ylab("GNI per capita, PPP (current international $)") + geom_point() + geom_path() + theme_bw() ################################################################## ## Choosing colors ################################################################## library(RColorBrewer) nCountries <- nlevels(CO2data$Country.Name) pal <- brewer.pal(n = 5, 'Set1') pal <- rep(pal, length = nCountries) ## Rank of average values of CO2 per capita CO2mean <- aggregate(CO2.capita ~ Country.Name, data = CO2data, FUN = mean) palOrdered <- pal[rank(CO2mean$CO2.capita)] library(reshape2) CO2capita <- CO2data[, c('Country.Name', 'Year', 'CO2.capita')] CO2capita <- dcast(CO2capita, Country.Name ~ Year) summary(CO2capita) hCO2 <- hclust(dist(CO2capita[, -1])) oldpar <- par(mar = c(0, 2, 0, 0) + .1) plot(hCO2, labels = CO2capita$Country.Name, xlab = '', ylab = '', sub = '', main = '') par(oldpar) idx <- match(levels(CO2data$Country.Name), CO2capita$Country.Name[hCO2$order]) palOrdered <- pal[idx] ## simpleTheme encapsulates the palette in a new theme for xyplot myTheme <- simpleTheme(pch = 19, cex = 0.6, col = palOrdered) ## lattice version pCO2.capita <- xyplot(GNI.capita ~ CO2.capita, data = CO2data, xlab = "Carbon dioxide emissions (metric tons per capita)", ylab = "GNI per capita, PPP (current international $)", groups = Country.Name, par.settings = myTheme, type = 'b') ## ggplot2 version gCO2.capita <- ggplot(data = CO2data, aes(x = CO2.capita, y = GNI.capita, color = Country.Name)) + geom_point() + geom_path() + scale_color_manual(values = palOrdered, guide = FALSE) + xlab('CO2 emissions (metric tons per capita)') + ylab('GNI per capita, PPP (current international $)') + theme_bw() ################################################################## ## Labels to show time information ################################################################## xyplot(GNI.capita ~ CO2.capita, data = CO2data, xlab = "Carbon dioxide emissions (metric tons per capita)", ylab = "GNI per capita, PPP (current international $)", groups = Country.Name, par.settings = myTheme, type = 'b', panel = function(x, y, ..., subscripts, groups){ panel.text(x, y, ..., labels = CO2data$Year[subscripts], pos = 2, cex = 0.5, col = 'gray') panel.superpose(x, y, subscripts, groups,...) }) ## lattice version pCO2.capita <- pCO2.capita + glayer_(panel.text(..., labels = CO2data$Year[subscripts], pos = 2, cex = 0.5, col = 'gray')) ## ggplot2 version gCO2.capita <- gCO2.capita + geom_text(aes(label = Year), colour = 'gray', size = 2.5, hjust = 0, vjust = 0) ################################################################## ## Country names: positioning labels ################################################################## library(directlabels) ## lattice version direct.label(pCO2.capita, method = 'extreme.grid') ## ggplot2 version direct.label(gCO2.capita, method = 'extreme.grid') ################################################################## ## A panel for each year ################################################################## ## lattice version xyplot(GNI.capita ~ CO2.capita | factor(Year), data = CO2data, xlab = "Carbon dioxide emissions (metric tons per capita)", ylab = "GNI per capita, PPP (current international $)", groups = Country.Name, type = 'b', auto.key = list(space = 'right')) ## ggplot2 version ggplot(data = CO2data, aes(x = CO2.capita, y = GNI.capita, colour = Country.Name)) + facet_wrap(~ Year) + geom_point(pch = 19) + xlab('CO2 emissions (metric tons per capita)') + ylab('GNI per capita, PPP (current international $)') + theme_bw() ## lattice version xyplot(GNI.capita ~ CO2.capita | factor(Year), data = CO2data, xlab = "Carbon dioxide emissions (metric tons per capita)", ylab = "GNI per capita, PPP (current international $)", groups = Country.Name, type = 'b', par.settings = myTheme) + glayer(panel.pointLabel(x, y, labels = group.value, col = palOrdered[group.number], cex = 0.7)) ################################################################## ## Using variable size to encode an additional variable ################################################################## library(classInt) z <- CO2data$CO2.PPP intervals <- classIntervals(z, n = 4, style = 'fisher') nInt <- length(intervals$brks) - 1 cex.key <- seq(0.5, 1.8, length = nInt) idx <- findCols(intervals) CO2data$cexPoints <- cex.key[idx] ggplot(data = CO2data, aes(x = CO2.capita, y = GNI.capita, colour = Country.Name)) + facet_wrap(~ Year) + geom_point(aes(size = cexPoints), pch = 19) + xlab('Carbon dioxide emissions (metric tons per capita)') + ylab('GNI per capita, PPP (current international $)') + theme_bw() op <- options(digits = 2) tab <- print(intervals) options(op) key <- list(space = 'right', title = expression(CO[2]/GNI.PPP), cex.title = 1, ## Labels of the key are the intervals strings text = list(labels = names(tab), cex = 0.85), ## Points sizes are defined with cex.key points = list(col = 'black', pch = 19, cex = cex.key, alpha = 0.7)) xyplot(GNI.capita ~ CO2.capita|factor(Year), data = CO2data, xlab = "Carbon dioxide emissions (metric tons per capita)", ylab = "GNI per capita, PPP (current international $)", groups = Country.Name, key = key, alpha = 0.7, panel = panel.superpose, panel.groups = function(x, y, subscripts, group.number, group.value, ...){ panel.xyplot(x, y, col = palOrdered[group.number], cex = CO2data$cexPoints[subscripts]) panel.pointLabel(x, y, labels = group.value, col = palOrdered[group.number], cex = 0.7) } ) library(plotly) p <- plot_ly(CO2data, x = ~CO2.capita, y = ~GNI.capita, sizes = c(10, 100), marker = list(opacity = 0.7, sizemode = 'diameter')) p <- add_markers(p, size = ~CO2.PPP, color = ~Country.Name, text = ~Country.Name, hoverinfo = "text", ids = ~Country.Name, frame = ~Year, showlegend = FALSE) p <- animation_opts(p, frame = 1000, transition = 800, redraw = FALSE) p <- animation_slider(p, currentvalue = list(prefix = "Year ")) p ################################################################## ## googleVis ################################################################## library(googleVis) pgvis <- gvisMotionChart(CO2data, xvar = 'CO2.capita', yvar = 'GNI.capita', sizevar = 'CO2.PPP', idvar = 'Country.Name', timevar = 'Year') print(pgvis, 'html', file = 'figs/googleVis.html') library(gridSVG) library(grid) xyplot(GNI.capita ~ CO2.capita, data = CO2data, xlab = "Carbon dioxide emissions (metric tons per capita)", ylab = "GNI per capita, PPP (current international $)", subset = Year==2000, groups = Country.Name, ## The limits of the graphic are defined ## with the entire dataset xlim = extendrange(CO2data$CO2.capita), ylim = extendrange(CO2data$GNI.capita), panel = function(x, y, ..., subscripts, groups) { color <- palOrdered[groups[subscripts]] radius <- CO2data$CO2.PPP[subscripts] ## Size of labels cex <- 1.1*sqrt(radius) ## Bubbles grid.circle(x, y, default.units = "native", r = radius*unit(.25, "inch"), name = trellis.grobname("points", type = "panel"), gp = gpar(col = color, ## Fill color ligther than border fill = adjustcolor(color, alpha = .5), lwd = 2)) ## Country labels grid.text(label = groups[subscripts], x = unit(x, 'native'), ## Labels above each bubble y = unit(y, 'native') + 1.5 * radius *unit(.25, 'inch'), name = trellis.grobname('labels', type = 'panel'), gp = gpar(col = color, cex = cex)) }) ## Duration in seconds of the animation duration <- 20 nCountries <- nlevels(CO2data$Country.Name) years <- unique(CO2data$Year) nYears <- length(years) ## Intermediate positions of the bubbles x_points <- animUnit(unit(CO2data$CO2.capita, 'native'), id = rep(seq_len(nCountries), each = nYears)) y_points <- animUnit(unit(CO2data$GNI.capita, 'native'), id = rep(seq_len(nCountries), each = nYears)) ## Intermediate positions of the labels y_labels <- animUnit(unit(CO2data$GNI.capita, 'native') + 1.5 * CO2data$CO2.PPP * unit(.25, 'inch'), id = rep(seq_len(nCountries), each = nYears)) ## Intermediate sizes of the bubbles size <- animUnit(CO2data$CO2.PPP * unit(.25, 'inch'), id = rep(seq_len(nCountries), each = nYears)) grid.animate(trellis.grobname("points", type = "panel", row = 1, col = 1), duration = duration, x = x_points, y = y_points, r = size, rep = TRUE) grid.animate(trellis.grobname("labels", type = "panel", row = 1, col = 1), duration = duration, x = x_points, y = y_labels, rep = TRUE) countries <- unique(CO2data$Country.Name) URL <- paste('http://en.wikipedia.org/wiki/', countries, sep = '') grid.hyperlink(trellis.grobname('points', type = 'panel', row = 1, col = 1), URL, group = FALSE) visibility <- matrix("hidden", nrow = nYears, ncol = nYears) diag(visibility) <- "visible" yearText <- animateGrob(garnishGrob(textGrob(years, .9, .15, name = "year", gp = gpar(cex = 2, col = "grey")), visibility = "hidden"), duration = 20, visibility = visibility, rep = TRUE) grid.draw(yearText) grid.export("figs/bubbles.svg")
Data
################################################################## ## SIAR ################################################################## ################################################################## ## Daily data of different meteorological variables ################################################################## library(zoo) aranjuez <- read.zoo("data/aranjuez.gz", index.column = 3, format = "%d/%m/%Y", fileEncoding = 'UTF-16LE', header = TRUE, fill = TRUE, sep = ';', dec = ",", as.is = TRUE) aranjuez <- aranjuez[, -c(1:4)] names(aranjuez) <- c('TempAvg', 'TempMax', 'TempMin', 'HumidAvg', 'HumidMax', 'WindAvg', 'WindMax', 'Radiation', 'Rain', 'ET') aranjuezClean <- within(as.data.frame(aranjuez),{ TempMin[TempMin > 40] <- NA HumidMax[HumidMax > 100] <- NA }) aranjuez <- zoo(aranjuezClean, index(aranjuez)) save(aranjuez, file = 'data/aranjuez.RData') ################################################################## ## Solar radiation measurements from different locations ################################################################## library(zoo) load('data/navarra.RData') ################################################################## ## Unemployment in the United States ################################################################## unemployUSA <- read.csv('data/unemployUSA.csv') nms <- unemployUSA$Series.ID ##columns of annual summaries annualCols <- 14 + 13*(0:12) ## Transpose. Remove annual summaries unemployUSA <- as.data.frame(t(unemployUSA[,-c(1, annualCols)])) ## First 7 characters can be suppressed names(unemployUSA) <- substring(nms, 7) library(zoo) Sys.setlocale("LC_TIME", 'C') idx <- as.yearmon(row.names(unemployUSA), format = '%b.%Y') unemployUSA <- zoo(unemployUSA, idx) unemployUSA <- unemployUSA[complete.cases(unemployUSA), ] save(unemployUSA, file = 'data/unemployUSA.RData') ################################################################## ## Gross National Income and $CO_2$ emissions ################################################################## library(WDI) CO2data <- WDI(indicator = c('EN.ATM.CO2E.PC', 'EN.ATM.CO2E.PP.GD', 'NY.GNP.MKTP.PP.CD', 'NY.GNP.PCAP.PP.CD'), start = 2000, end = 2014, country = c('BR', 'CN', 'DE', 'ES', 'FI', 'FR', 'GR', 'IN', 'NO', 'US')) names(CO2data) <- c('iso2c', 'Country.Name', 'Year', 'CO2.capita', 'CO2.PPP', 'GNI.PPP', 'GNI.capita') CO2data <- CO2data[complete.cases(CO2data), ] CO2data$Country.Name <- factor(CO2data$Country.Name) save(CO2data, file = 'data/CO2.RData')